d3pmga1d1e8ca1
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
3pmg_Ah
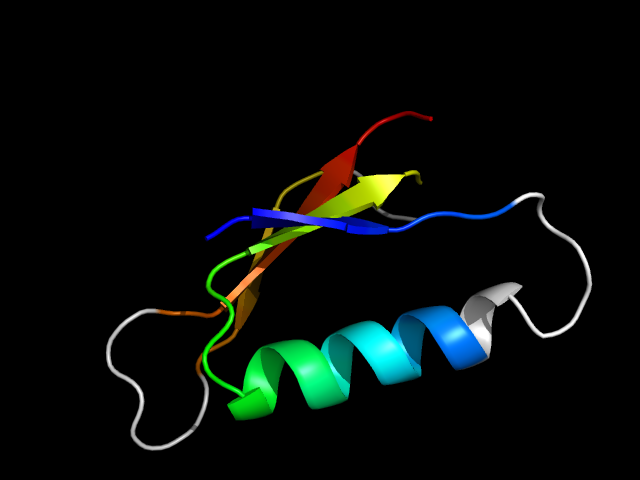 |
d3pmga1 | A54-A89,A167-A190 | Alpha and beta proteins (a/b) | Phosphoglucomutase, first 3 domains | Phosphoglucomutase, first 3 domains | Phosphoglucomutase, first 3 domains | Phosphoglucomutase |
1e8c_Ac
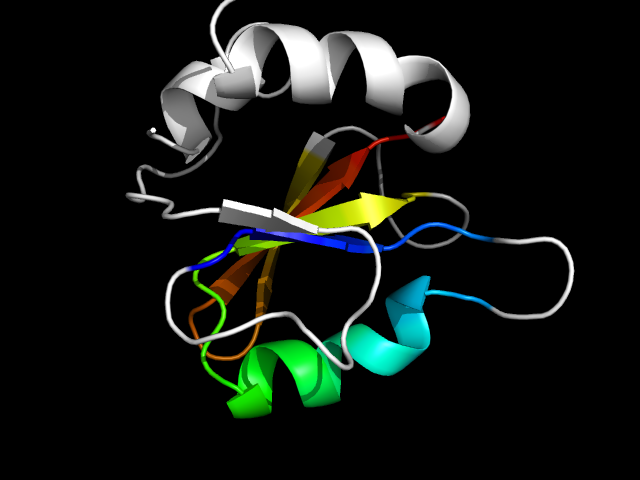 |
d1e8ca1 | A3-A67,A68-A103 | Alpha and beta proteins (a/b) | MurF and HprK N-domain-like | MurE/MurF N-terminal domain | MurE/MurF N-terminal domain | UDP-N-acetylmuramyl tripeptide synthetase MurE
|
Justification for analogy
The motif in 3pmg: comparing the three duplicates, the C-terminal beta hairpin in the first duplicate is an addition. Hybrid motif. These
three duplicates apparently have a mini-Rossmann core.
The motif in 1e8c: compare 1e8c_A with d1gg4a3 (same family), it appears that the antiparallel strand in 1e8c_A is originally a helix or loop
connecting the two parallel strands. Anyway, this can be regarded as a core motif.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
3pmg_Ah --------------------------------ATLVVGGdgrfymkeAIQLIVRIAAANGIGRLVIGQ----gvlGKQQFDlenkfkPFTVEIVDS---------------
1e8c_Ac rnlrdllapwvpdapsralremtldsrvaaagDLFVAVVgh----qaDGRRYIPQAIAQGVAAIIAEAkdeatdgEIREMH------GVPVIYLSQlnerlsalagrfyhe
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
3pmg_Ah --------------------------------ATLVVGGDgrFYMKeaIQLIVRIAAANGIGRLVIGQ----GVLGKQQFDlenkfkPFTVEIVDS---------------
1e8c_Ac rnlrdllapwvpdapsralremtldsrvaaagDLFVAVVG--HQAD--GRRYIPQAIAQGVAAIIAEAkdeaTDGEIREMH------GVPVIYLSQlnerlsalagrfyhe
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
3pmg_Ah --------------------------------ATLVVGGDGRFYMKEAIQLIVRIAAANGIGRLVIGQ--GVL--GKQQFDLENKFKPFTVEIVDS---------------
1e8c_Ac RNLRDLLAPWVPDAPSRALREMTLDSRVAAAGDLFVAVVG-HQ-AD-GRR-YIPQAIAQGVAAIIAEAKDEATDGEIREMH------GVPVIYLSQLNERLSALAGRFYHE
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
3pmg_Ah --------------------------------ATLVVGGDG-RFYMKEAIQLIVRIAAANGIGRLVIGQGV-------LGKQQFDLENKFKP-FTVEIVDS----------------
1e8c_Ac RNLRDLLAPWVPDAPSRALREMTLDSRVAAAGDLFVAVV--GHQAD---GRRYIPQAIAQGVAAIIAE---AKDEATDGEIREMH-------GVPVIYLS-QLNERLSALAGRFYHE

