d6mhta_d1vm0b_
Motif
(click icon to enlarge) | SCOPid | Range | SCOP Class | SCOP Fold | SCOP Superfamily | SCOP Family | Protein |
|---|
6mht_Ah
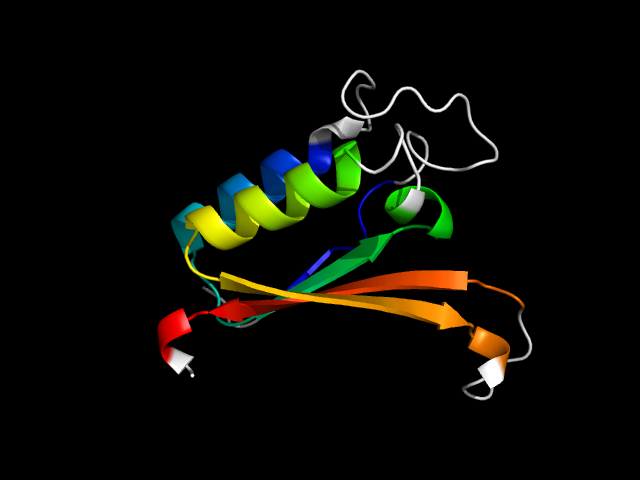 |
d6mhta_ | A71-A176 | Alpha and beta proteins (a/b) | S-adenosyl-L-methionine-dependent methyltransferases | S-adenosyl-L-methionine-dependent methyltransferases | C5 cytosine-specific DNA methylase, DCM | DNA methylase HhaI |
1vm0_Bc
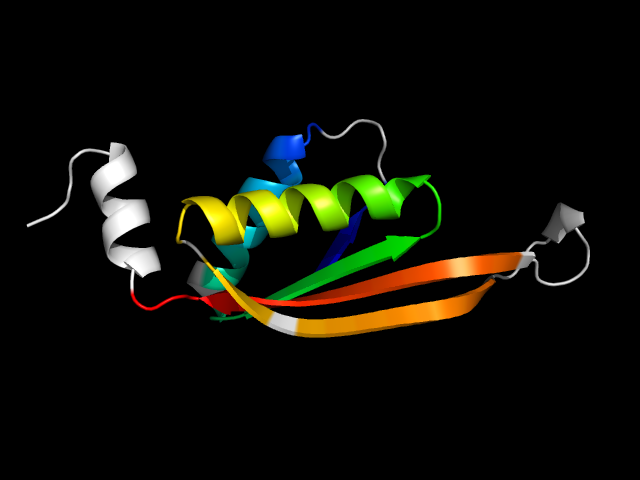 |
d1vm0b_ | B20-B122 | Alpha and beta proteins (a+b) | IF3-like | AlbA-like | Hypothetical protein At2g34160 | Hypothetical protein At2g34160
|
Justification for analogy
The motif in 6mht: methyltransferase is homologous to Rossmann-like proteins, so the anti-parallel strand is an insertion. So the motif is
analogous to IF3-like.
In the alignments below, two upper-case letters are aligned, while two lower-case letters are NOT aligned. The two sequences in an alignment are NOT continuous in most cases (see their ranges). The superpositions are re-constructed from the corresponding alignments by minimizing the overall RMSD. In a PDB-format superposition file, the first domain is represented by chain S and the second domain is represented by chain A.
MANUAL alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
6mht_Ah dhdiLCAGFPcqafsisgkqkgfedsrgtLFFDIARIVREKK--PKVVFMENVKNFAshdngNTLEVVKNTMNELD-YSF-HAKVLNALdyg----ipqKRERIYMICFRNDln------------
1vm0_Bc ---kNRIQVSnt---------------kkPLFFYVNLAKRYMqqYNDVELSALGMAI-----ATVVTVTEILKNNGfAVEkKIMTSIVDikddargrpvQKAKIEITLVKSEkfdelmaaaneeke
DALI alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
6mht_Ah dhdILCAGFPCQafsisgkqkgfedSRGT-LFFDIARIVreKKPKVVFMENVKNFAshdngnTLEVVKNTMNE-LDYSF-HAKVLNAL----dygiPQKRERIYMICFRND------------ln
1vm0_Bc ---KNRIQVSNT-------------KKPLfFYVNLAKRY-mQQYNDVELSALGMAI-----aTVVTVTEILKNnGFAVEkKIMTSIVDikddargrPVQKAKIEITLVKSEkfdelmaaaneeke
TM alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
6mht_Ah DHDILCAGFPCQAFSISGKQKGFEDSRGT--LFFDIARIVREKKPKVVFMENVKNFASHDNGNTLEVVKNTMN--ELDYSF-HAKVLNAL-------DYGIPQ-KRERIYMICFRND--------------LN
1vm0_Bc ---KNRIQV-SNT-------------KKPLFFYVNLAKRYMQQ-YNDVELSALGMAIAT----VV-TVTEILKNNGF-AVEKKIMTSIVDIKDDARG----RPVQKAKIEITLVKSEKFDELMAAANEEKE--
FAST alignment PDB format superposition aligned motifs in PyMol whole structures in PyMol
6mht_Ah DHDI-LCAGFPCQAFSISGKQKGFEDSRGTL------FFDIARIVREKK---PKVVFMENVKNFASHDNGNTLEVVKNTMNELD-YSF-HAKVLNALDYGIP---------QKRERIYMICFRNDLN--------------
1vm0_Bc ----KNRIQV---------------------SNTKKPLFFYVNLAKRY-MQQYNDVELSALGMAI-----ATVVTVTEILKNNGFAVEKKIMTSIVD-----IKDDARGRPVQKAKIEITLVKSE--KFDELMAAANEEKE

