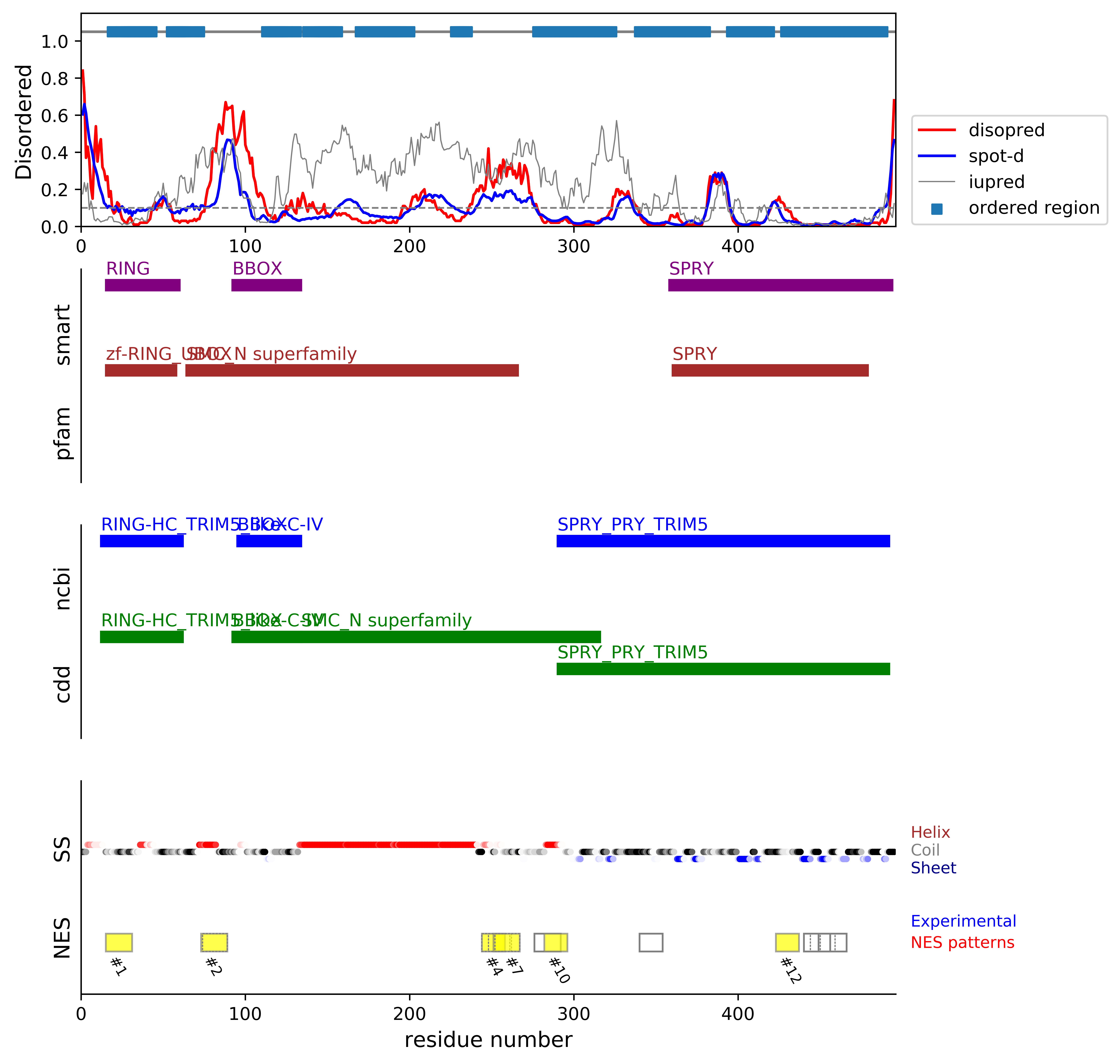

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | A1E965 | 15 | CPICLELLTEPLSLHC | CCCCCCCCCCCCCCCC | c1aR-4 | uniq | 0.102 | 0.096 | 0.027 | boundary | boundary|RING-HC_TRIM5_like-C-IV; | 0.0 |
| 2 | fp_D | A1E965 | 73 | HVANIVEKLREVKLSP | HHHHHHHHHHCCCCCC | c1a-5 | multi-selected | 0.351 | 0.184 | 0.378 | boundary | boundary|RING-HC_TRIM5_like-C-IV; | 0.0 |
| 3 | fp_D | A1E965 | 74 | VANIVEKLREVKLSP | HHHHHHHHHCCCCCC | c1b-4 | multi | 0.37 | 0.188 | 0.376 | boundary | boundary|RING-HC_TRIM5_like-C-IV; | 0.0 |
| 4 | fp_D | A1E965 | 244 | SMMDLLQGVDGIIK | CHHHHHCCCHHHHC | c3-4 | multi-selected | 0.269 | 0.161 | 0.288 | boundary | MID|SMC_N superfamily; | 0.0 |
| 5 | fp_D | A1E965 | 244 | SMMDLLQGVDGIIKRIE | CHHHHHCCCHHHHCCCC | c1cR-5 | multi | 0.276 | 0.165 | 0.295 | boundary | MID|SMC_N superfamily; | 0.0 |
| 6 | fp_D | A1E965 | 248 | LLQGVDGIIKRIEN | HHCCCHHHHCCCCC | c3-4 | multi | 0.294 | 0.168 | 0.291 | boundary | MID|SMC_N superfamily; | 0.0 |
| 7 | fp_D | A1E965 | 251 | GVDGIIKRIENMTLKK | CCHHHHCCCCCCCCCC | c1a-5 | multi-selected | 0.291 | 0.171 | 0.323 | __ | MID|SMC_N superfamily; | 0.0 |
| 8 | fp_D | A1E965 | 252 | VDGIIKRIENMTLKK | CHHHHCCCCCCCCCC | c1b-4 | multi | 0.296 | 0.172 | 0.327 | boundary | MID|SMC_N superfamily; | 0.0 |
| 9 | fp_D | A1E965 | 276 | RVFRAPDLKGMLDMFR | CCCCCCCHHHHHHHHH | c1d-AT-5 | uniq | 0.044 | 0.052 | 0.279 | boundary | MID|SMC_N superfamily; boundary|SPRY_PRY_TRIM5; | 0.0 |
| 10 | fp_D | A1E965 | 282 | DLKGMLDMFRELTD | CHHHHHHHHHHHCC | c3-4 | uniq | 0.026 | 0.038 | 0.214 | boundary | boundary|SMC_N superfamily; boundary|SPRY_PRY_TRIM5; | 0.0 |
| 11 | fp_D | A1E965 | 340 | SLTNFNYCTGVLGS | CCCCCCCCCEEECC | c3-AT | uniq | 0.019 | 0.03 | 0.114 | boundary | boundary|SMC_N superfamily; MID|SPRY_PRY_TRIM5; | 0.143 |
| 12 | fp_D | A1E965 | 423 | PFAPFIVPLSVIIC | CCCCCCCCCCCCCC | c3-4 | uniq | 0.094 | 0.05 | 0.019 | boundary | MID|SPRY_PRY_TRIM5; | 0.0 |
| 13 | fp_D | A1E965 | 440 | VGVFVDYEACTVSFFN | EEEEECCCCCEEEEEE | c1a-AT-4 | multi-selected | 0.01 | 0.01 | 0.013 | boundary | MID|SPRY_PRY_TRIM5; | 0.286 |
| 14 | fp_beta_O | A1E965 | 444 | VDYEACTVSFFNITN | ECCCCCEEEEEECCC | c1b-AT-4 | multi | 0.01 | 0.012 | 0.012 | ORD | MID|SPRY_PRY_TRIM5; | 0.714 |
| 15 | fp_O | A1E965 | 449 | CTVSFFNITNHGFLIYK | CEEEEEECCCCCCCEEE | c1c-AT-4 | multi-selected | 0.01 | 0.015 | 0.01 | ORD | MID|SPRY_PRY_TRIM5; | 0.375 |
| 16 | fp_O | A1E965 | 450 | TVSFFNITNHGFLIYK | EEEEEECCCCCCCEEE | c1d-AT-5 | multi | 0.01 | 0.015 | 0.01 | ORD | MID|SPRY_PRY_TRIM5; | 0.286 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment