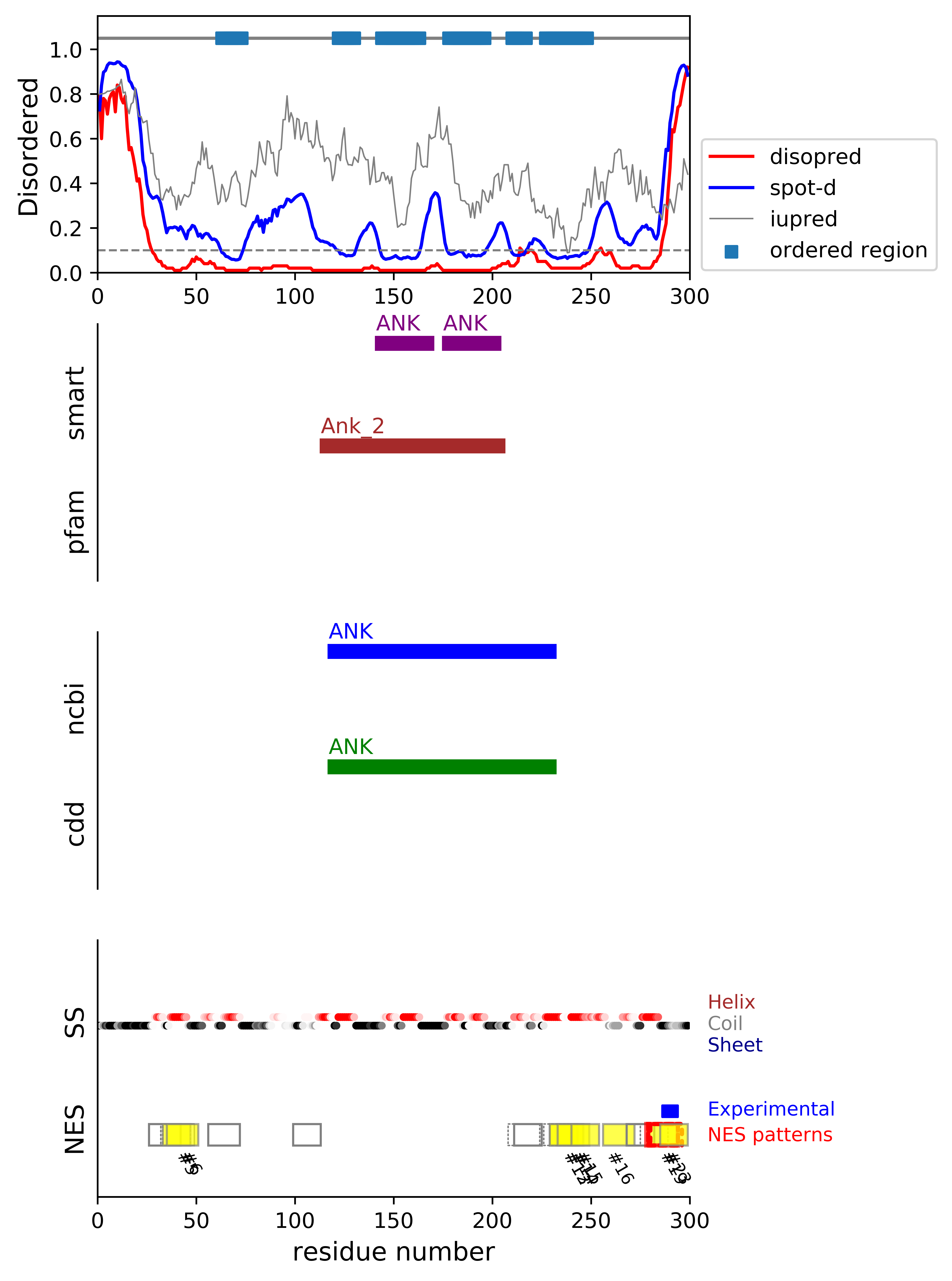

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | A7YB12 | 26 | APEPLAEAGGLVSFAD | CCCHHHHHCCCHHHHH | c1d-AT-4 | uniq | 0.047 | 0.264 | 0.399 | DISO | 0.0 | |
| 2 | fp_D | A7YB12 | 32 | EAGGLVSFADFGVSLGS | HHCCCHHHHHHHHHCCC | c1c-AT-4 | multi | 0.025 | 0.205 | 0.345 | boundary | 0.0 | |
| 3 | fp_D | A7YB12 | 32 | EAGGLVSFADFGVSL | HHCCCHHHHHHHHHC | c1b-4 | multi | 0.021 | 0.206 | 0.34 | DISO | 0.0 | |
| 4 | fp_D | A7YB12 | 33 | AGGLVSFADFGVSLGS | HCCCHHHHHHHHHCCC | c1d-AT-4 | multi | 0.023 | 0.199 | 0.346 | boundary | 0.0 | |
| 5 | fp_D | A7YB12 | 33 | AGGLVSFADFGVSL | HCCCHHHHHHHHHC | c2-4 | multi-selected | 0.019 | 0.198 | 0.341 | DISO | 0.0 | |
| 6 | fp_D | A7YB12 | 35 | GLVSFADFGVSLGSGA | CCHHHHHHHHHCCCCC | c1aR-4 | multi-selected | 0.027 | 0.193 | 0.357 | boundary | 0.0 | |
| 7 | fp_D | A7YB12 | 56 | SVGRAQSSLRYLQVLW | HHHHHCCCHHHHHHHH | c1a-AT-5 | uniq | 0.021 | 0.102 | 0.397 | boundary | 0.0 | |
| 8 | fp_D | A7YB12 | 99 | RLGPTGKEVHALKR | CCCCCCHHHCCCCH | c3-AT | uniq | 0.017 | 0.28 | 0.63 | boundary | boundary|ANK; | 0.0 |
| 9 | fp_D | A7YB12 | 208 | GRTPLHLAKSKLNILQ | CCCHHHHHHHCCCHHH | c1d-AT-4 | multi | 0.07 | 0.108 | 0.389 | boundary | boundary|ANK; | 0.0 |
| 10 | fp_D | A7YB12 | 211 | PLHLAKSKLNILQE | HHHHHHHCCCHHHC | c3-AT | multi-selected | 0.076 | 0.109 | 0.368 | boundary | boundary|ANK; | 0.0 |
| 11 | fp_D | A7YB12 | 226 | HSQCLEAVRLEVKQ | CHHHHHHHHHHHHH | c2-AT-4 | multi | 0.026 | 0.076 | 0.207 | boundary | boundary|ANK; | 0.0 |
| 12 | fp_D | A7YB12 | 229 | CLEAVRLEVKQIIH | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.021 | 0.07 | 0.183 | boundary | boundary|ANK; | 0.0 |
| 13 | fp_D | A7YB12 | 229 | CLEAVRLEVKQIIHMLR | HHHHHHHHHHHHHHHHH | c1c-5 | multi-selected | 0.021 | 0.071 | 0.188 | boundary | boundary|ANK; | 0.0 |
| 14 | fp_D | A7YB12 | 233 | VRLEVKQIIHMLREYL | HHHHHHHHHHHHHHHH | c1aR-4 | multi-selected | 0.022 | 0.074 | 0.205 | boundary | boundary|ANK; | 0.0 |
| 15 | fp_D | A7YB12 | 240 | IIHMLREYLERLGR | HHHHHHHHHHHHHH | c3-4 | uniq | 0.039 | 0.112 | 0.284 | boundary | boundary|ANK; | 0.0 |
| 16 | fp_D | A7YB12 | 256 | QRERLDDLCTRLQMTS | HHCCCCCCCHHHHHHH | c1d-4 | uniq | 0.048 | 0.206 | 0.459 | boundary | boundary|ANK; | 0.0 |
| 17 | fp_D | A7YB12 | 268 | QMTSTKEQVDEVTD | HHHHHCCCHHHHHH | c3-AT | uniq | 0.024 | 0.173 | 0.389 | DISO | 0.0 | |
| 18 | fp_D | A7YB12 | 275 | QVDEVTDLLASFTS | CHHHHHHHHHCCCC | c3-4 | multi | 0.067 | 0.25 | 0.325 | DISO | 0.0 | |
| 19 | cand_D | A7YB12 | 278 | EVTDLLASFTSLSLQM * * | HHHHHHHCCCCCCCCC | c1a-5 | multi-selected | 0.229 | 0.405 | 0.306 | DISO | 0.0 | |
| 20 | cand_D | A7YB12 | 279 | VTDLLASFTSLSLQM * * | HHHHHHCCCCCCCCC | c1b-4 | multi | 0.243 | 0.418 | 0.297 | DISO | 0.0 | |
| 21 | cand_D | A7YB12 | 279 | VTDLLASFTSLSLQMQS * * | HHHHHHCCCCCCCCCCC | c1c-AT-4 | multi | 0.302 | 0.475 | 0.309 | DISO | 0.0 | |
| 22 | cand_D | A7YB12 | 280 | TDLLASFTSLSLQM * * | HHHHHCCCCCCCCC | c2-AT-4 | multi | 0.259 | 0.434 | 0.294 | DISO | 0.0 | |
| 23 | fp_D | A7YB12 | 285 | SFTSLSLQMQSMEK * * | CCCCCCCCCCCCCC | c3-4 | multi-selected | 0.531 | 0.698 | 0.34 | DISO | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment