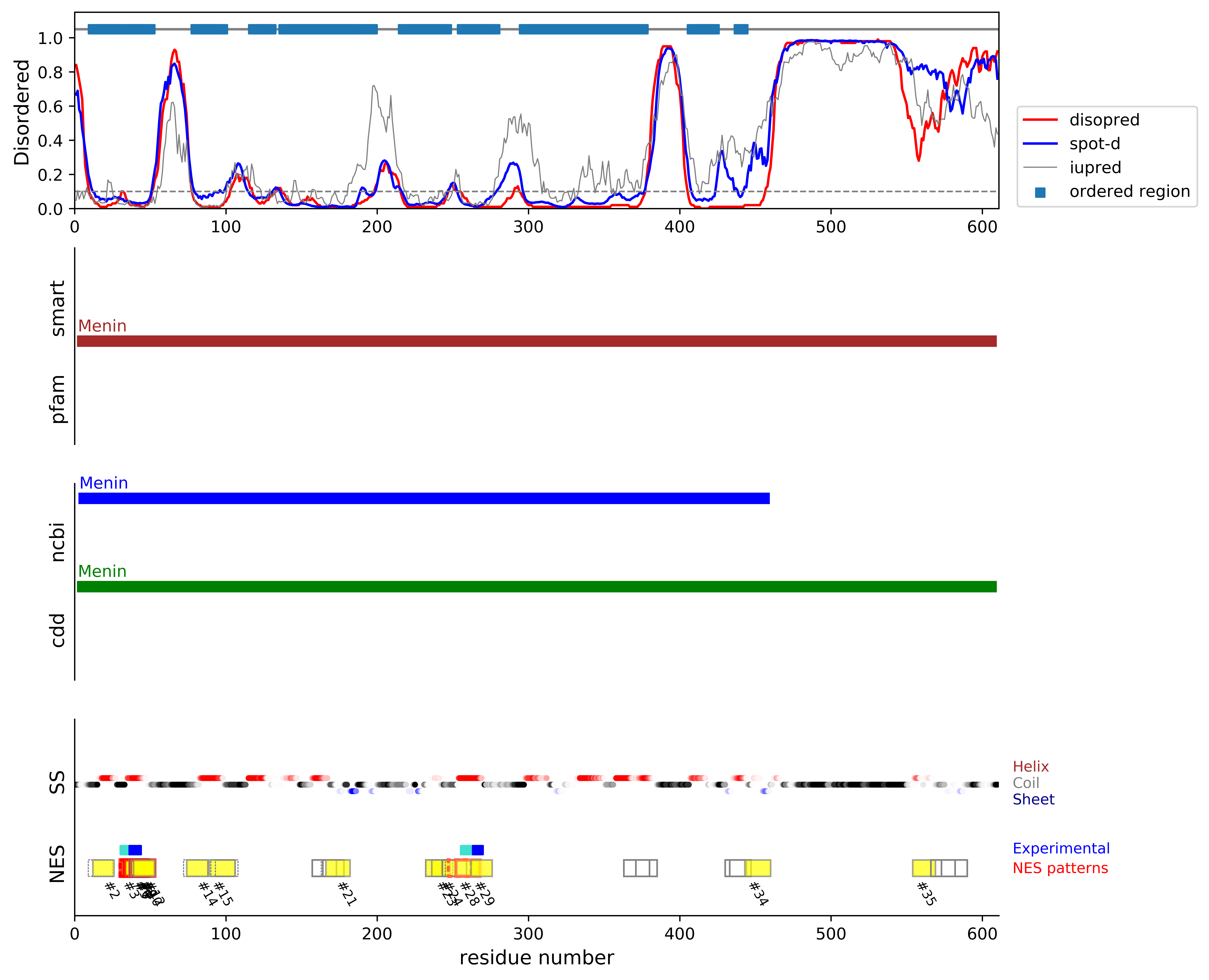

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | O00255 | 9 | TLFPLRSIDDVVRLFA | CCCCCCCHHHHHHHHH | c1d-5 | multi | 0.035 | 0.082 | 0.071 | boundary | boundary|Menin; | 0.0 |
| 2 | fp_D | O00255 | 12 | PLRSIDDVVRLFAA | CCCCHHHHHHHHHH | c3-4 | multi-selected | 0.023 | 0.063 | 0.071 | boundary | boundary|Menin; | 0.0 |
| 3 | cand_D | O00255 | 30 | EEPDLVLLSLVLGF ++++++*+* | CCCCHHHHHHHHHH | c2-4 | multi-selected | 0.044 | 0.041 | 0.023 | boundary | MID|Menin; | 0.0 |
| 4 | cand_D | O00255 | 30 | EEPDLVLLSLVLGFVE ++++++*+* | CCCCHHHHHHHHHHHH | c1d-4 | multi | 0.04 | 0.04 | 0.021 | boundary | MID|Menin; | 0.0 |
| 5 | cand_D | O00255 | 32 | PDLVLLSLVLGFVEHF ++++++*+* | CCHHHHHHHHHHHHHH | c1aR-4 | multi-selected | 0.032 | 0.037 | 0.019 | boundary | MID|Menin; | 0.0 |
| 6 | cand_D | O00255 | 33 | DLVLLSLVLGFVEHFL ++++++*+* | CHHHHHHHHHHHHHHC | c1aR-5 | multi-selected | 0.027 | 0.036 | 0.018 | boundary | MID|Menin; | 0.0 |
| 7 | cand_D | O00255 | 33 | DLVLLSLVLGFVEH ++++++*+* | CHHHHHHHHHHHHH | c3-4 | multi-selected | 0.028 | 0.036 | 0.019 | boundary | MID|Menin; | 0.0 |
| 8 | fp_D | O00255 | 36 | LLSLVLGFVEHFLA +++*+* | HHHHHHHHHHHHCC | c3-4 | multi-selected | 0.019 | 0.036 | 0.016 | boundary | MID|Menin; | 0.0 |
| 9 | fp_D | O00255 | 36 | LLSLVLGFVEHFLAVNR +++*+* | HHHHHHHHHHHHCCCCC | c1c-5 | multi-selected | 0.031 | 0.05 | 0.022 | boundary | MID|Menin; | 0.0 |
| 10 | cand_D | O00255 | 37 | LSLVLGFVEHFLAVNR ++*+* | HHHHHHHHHHHCCCCC | c1aR-4 | multi-selected | 0.031 | 0.05 | 0.02 | boundary | MID|Menin; | 0.0 |
| 11 | fp_D | O00255 | 37 | LSLVLGFVEHFLAVNR ++*+* | HHHHHHHHHHHCCCCC | c1d-4 | multi | 0.031 | 0.05 | 0.02 | boundary | MID|Menin; | 0.0 |
| 12 | fp_D | O00255 | 39 | LVLGFVEHFLAVNR *+* | HHHHHHHHHCCCCC | c3-4 | multi-selected | 0.033 | 0.052 | 0.02 | boundary | MID|Menin; | 0.0 |
| 13 | fp_D | O00255 | 72 | PGGLTYFPVADLSIIA | CCCCCCCCCCCHHHHH | c1a-AT-4 | multi | 0.184 | 0.207 | 0.118 | boundary | MID|Menin; | 0.0 |
| 14 | fp_D | O00255 | 74 | GLTYFPVADLSIIA | CCCCCCCCCHHHHH | c2-5 | multi-selected | 0.114 | 0.149 | 0.088 | boundary | MID|Menin; | 0.0 |
| 15 | fp_D | O00255 | 89 | LYARFTAQIRGAVDLSL | HHHHHHHHHHCCCCCCC | c1c-4 | multi-selected | 0.042 | 0.119 | 0.067 | boundary | MID|Menin; | 0.0 |
| 16 | fp_D | O00255 | 90 | YARFTAQIRGAVDLSL | HHHHHHHHHCCCCCCC | c1d-AT-4 | multi | 0.044 | 0.121 | 0.07 | boundary | MID|Menin; | 0.0 |
| 17 | fp_D | O00255 | 93 | FTAQIRGAVDLSLYP | HHHHHHCCCCCCCCC | c1b-AT-4 | multi | 0.07 | 0.144 | 0.101 | boundary | MID|Menin; | 0.0 |
| 18 | fp_O | O00255 | 157 | VAFAVVGACQALGLRD | HHHHHHHHHHHCCCCC | c1d-AT-4 | uniq | 0.02 | 0.013 | 0.063 | ORD | MID|Menin; | 0.0 |
| 19 | fp_O | O00255 | 163 | GACQALGLRDVHLAL | HHHHHCCCCCCEEEE | c1b-AT-4 | multi | 0.01 | 0.012 | 0.048 | ORD | MID|Menin; | 0.0 |
| 20 | fp_O | O00255 | 164 | ACQALGLRDVHLAL | HHHHCCCCCCEEEE | c2-4 | multi-selected | 0.01 | 0.013 | 0.047 | ORD | MID|Menin; | 0.0 |
| 21 | fp_D | O00255 | 166 | QALGLRDVHLALSEDH | HHCCCCCCEEEECCCE | c1aR-4 | multi-selected | 0.01 | 0.013 | 0.056 | boundary | MID|Menin; | 0.429 |
| 22 | fp_D | O00255 | 232 | RKMEVAFMVCAINPSIDL | CCCCHHHHHHCCCCCCCC | c4-4 | multi | 0.038 | 0.056 | 0.066 | boundary | MID|Menin; | 0.0 |
| 23 | fp_D | O00255 | 232 | RKMEVAFMVCAINPSI | CCCCHHHHHHCCCCCC | c1aR-4 | multi-selected | 0.027 | 0.046 | 0.065 | boundary | MID|Menin; | 0.0 |
| 24 | fp_D | O00255 | 236 | VAFMVCAINPSIDLHT | HHHHHHCCCCCCCCCC | c1aR-4 | multi-selected | 0.056 | 0.073 | 0.079 | boundary | MID|Menin; | 0.0 |
| 25 | fp_D | O00255 | 243 | INPSIDLHTDSLELLQ + | CCCCCCCCCCCHHHHH | c1a-AT-4 | multi-selected | 0.088 | 0.086 | 0.1 | boundary | MID|Menin; | 0.0 |
| 26 | fp_D | O00255 | 245 | PSIDLHTDSLELLQ + | CCCCCCCCCHHHHH | c2-AT-4 | multi | 0.096 | 0.091 | 0.102 | boundary | MID|Menin; | 0.0 |
| 27 | cand_D | O00255 | 247 | IDLHTDSLELLQLQQ ++++ | CCCCCCCHHHHHHHH | c1b-AT-4 | multi | 0.091 | 0.078 | 0.092 | boundary | MID|Menin; | 0.0 |
| 28 | cand_D | O00255 | 252 | DSLELLQLQQKLLWLL ++++++++** | CCHHHHHHHHHHHHHH | c1aR-5 | multi-selected | 0.053 | 0.036 | 0.068 | boundary | MID|Menin; | 0.0 |
| 29 | fp_D | O00255 | 262 | KLLWLLYDLGHLER ++++** | HHHHHHHHCCCCCC | c3-4 | uniq | 0.011 | 0.03 | 0.029 | boundary | MID|Menin; | 0.0 |
| 30 | fp_D | O00255 | 363 | EFFEVANDVIPNLLKEA | HHHHHHHHHHHHHHHHH | c1cR-4 | uniq | 0.048 | 0.084 | 0.196 | boundary | MID|Menin; | 0.0 |
| 31 | fp_D | O00255 | 371 | VIPNLLKEAASLLE | HHHHHHHHHHHHHH | c3-AT | uniq | 0.257 | 0.198 | 0.31 | boundary | MID|Menin; | 0.0 |
| 32 | fp_D | O00255 | 430 | PVLHVGWATFLVQS | CEEEEHHHHHHHHH | c3-AT | multi-selected | 0.011 | 0.141 | 0.343 | boundary | MID|Menin; | 0.286 |
| 33 | fp_D | O00255 | 433 | HVGWATFLVQSLGR | EEHHHHHHHHHHCC | c3-AT | multi-selected | 0.013 | 0.135 | 0.346 | boundary | MID|Menin; | 0.0 |
| 34 | fp_D | O00255 | 443 | SLGRFEGQVRQKVRIVS | HHCCCCHHHHHEEEEEE | c1c-5 | uniq | 0.049 | 0.284 | 0.433 | DISO | MID|Menin; | 0.125 |
| 35 | fp_D | O00255 | 554 | QSEKMKGMKELLVAT | CHHHHHCCHHHHHHC | c1b-4 | uniq | 0.441 | 0.816 | 0.619 | DISO | MID|Menin; | 0.0 |
| 36 | fp_D | O00255 | 566 | VATKINSSAIKLQLTA | HHCCCCCCCCEEEEEC | c1a-AT-4 | uniq | 0.626 | 0.728 | 0.57 | DISO | MID|Menin; | 0.143 |
| 37 | fp_beta_D | O00255 | 573 | SAIKLQLTAQSQVQMKK | CCCEEEEECCCEEEECC | c1c-AT-4 | uniq | 0.763 | 0.66 | 0.649 | DISO | boundary|Menin; | 0.625 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment