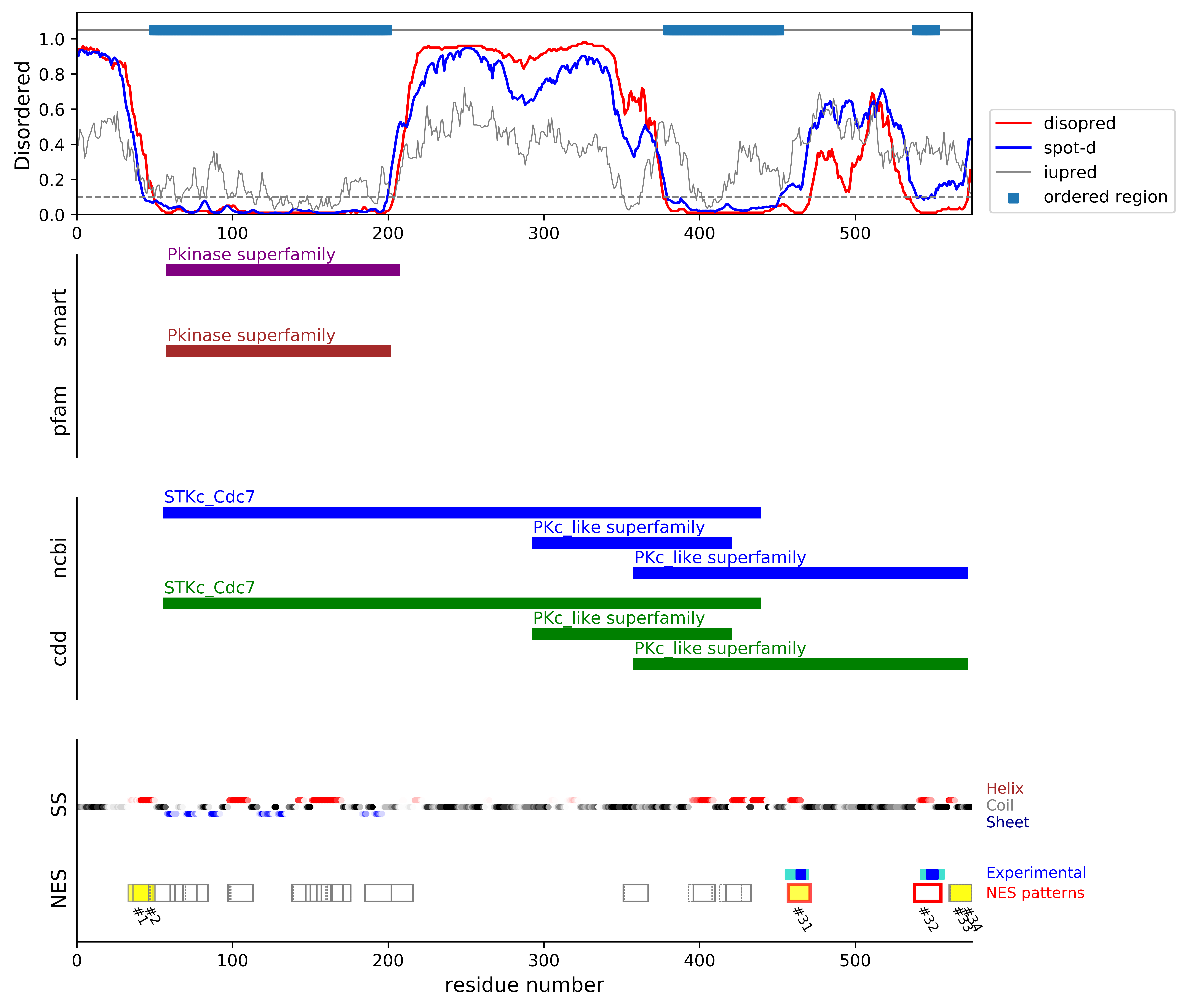

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | O00311 | 33 | QNFKLAGVKKDIEKLY | CHHHHHHHHHHHHHHH | c1aR-5 | multi-selected | 0.432 | 0.254 | 0.242 | boundary | boundary|STKc_Cdc7; | 0.0 |
| 2 | fp_D | O00311 | 36 | KLAGVKKDIEKLYE | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.351 | 0.183 | 0.206 | boundary | boundary|STKc_Cdc7; | 0.0 |
| 3 | fp_D | O00311 | 46 | KLYEAVPQLSNVFKIED | HHHHHCCCCCCCEEEEE | c1c-AT-5 | multi-selected | 0.058 | 0.058 | 0.167 | boundary | boundary|STKc_Cdc7; | 0.0 |
| 4 | fp_D | O00311 | 46 | KLYEAVPQLSNVFK | HHHHHCCCCCCCEE | c3-AT | multi | 0.067 | 0.062 | 0.159 | boundary | boundary|STKc_Cdc7; | 0.0 |
| 5 | fp_D | O00311 | 47 | LYEAVPQLSNVFKIED | HHHHCCCCCCCEEEEE | c1d-4 | multi | 0.052 | 0.057 | 0.164 | boundary | boundary|STKc_Cdc7; | 0.0 |
| 6 | fp_beta_D | O00311 | 60 | IEDKIGEGTFSSVYLAT | EEEEECCCCEEEEEEEE | c1c-AT-4 | uniq | 0.019 | 0.028 | 0.152 | boundary | boundary|STKc_Cdc7; | 0.5 |
| 7 | fp_beta_O | O00311 | 68 | TFSSVYLATAQLQVGP | CEEEEEEEEEECCCCC | c1a-AT-5 | multi-selected | 0.017 | 0.032 | 0.151 | ORD | boundary|STKc_Cdc7; | 1.0 |
| 8 | fp_beta_O | O00311 | 68 | TFSSVYLATAQLQVGP | CEEEEEEEEEECCCCC | c1d-AT-5 | multi | 0.017 | 0.032 | 0.151 | ORD | boundary|STKc_Cdc7; | 1.0 |
| 9 | fp_beta_O | O00311 | 70 | SSVYLATAQLQVGP | EEEEEEEEECCCCC | c2-AT-4 | multi | 0.017 | 0.032 | 0.155 | ORD | boundary|STKc_Cdc7; | 0.857 |
| 10 | fp_O | O00311 | 97 | HPIRIAAELQCLTVAG | CHHHHHHHHHHHHHCC | c1a-4 | multi-selected | 0.028 | 0.019 | 0.146 | ORD | MID|STKc_Cdc7; | 0.0 |
| 11 | fp_O | O00311 | 98 | PIRIAAELQCLTVAG | HHHHHHHHHHHHHCC | c1b-AT-5 | multi | 0.027 | 0.017 | 0.149 | ORD | MID|STKc_Cdc7; | 0.0 |
| 12 | fp_O | O00311 | 99 | IRIAAELQCLTVAG | HHHHHHHHHHHHCC | c2-AT-4 | multi | 0.025 | 0.016 | 0.153 | ORD | MID|STKc_Cdc7; | 0.0 |
| 13 | fp_O | O00311 | 138 | EHESFLDILNSLSFQE | CCCCHHHHHCCCCHHH | c1a-4 | multi-selected | 0.01 | 0.022 | 0.056 | ORD | MID|STKc_Cdc7; | 0.0 |
| 14 | fp_O | O00311 | 138 | EHESFLDILNSLSFQE | CCCCHHHHHCCCCHHH | c1d-4 | multi | 0.01 | 0.022 | 0.056 | ORD | MID|STKc_Cdc7; | 0.0 |
| 15 | fp_O | O00311 | 139 | HESFLDILNSLSFQE | CCCHHHHHCCCCHHH | c1b-4 | multi | 0.01 | 0.022 | 0.055 | ORD | MID|STKc_Cdc7; | 0.0 |
| 16 | fp_O | O00311 | 147 | NSLSFQEVREYMLNLF | CCCCHHHHHHHHHHHH | c1aR-5 | multi-selected | 0.01 | 0.013 | 0.06 | ORD | MID|STKc_Cdc7; | 0.0 |
| 17 | fp_O | O00311 | 150 | SFQEVREYMLNLFK | CHHHHHHHHHHHHH | c3-4 | multi-selected | 0.01 | 0.01 | 0.055 | ORD | MID|STKc_Cdc7; | 0.0 |
| 18 | fp_O | O00311 | 157 | YMLNLFKALKRIHQ | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.01 | 0.009 | 0.067 | ORD | MID|STKc_Cdc7; | 0.0 |
| 19 | fp_O | O00311 | 160 | NLFKALKRIHQFGIVH | HHHHHHHHHHHCCCCC | c1a-AT-5 | multi | 0.012 | 0.012 | 0.096 | ORD | MID|STKc_Cdc7; | 0.0 |
| 20 | fp_O | O00311 | 161 | LFKALKRIHQFGIVH | HHHHHHHHHHCCCCC | c1b-5 | multi | 0.012 | 0.013 | 0.097 | ORD | MID|STKc_Cdc7; | 0.0 |
| 21 | fp_beta_D | O00311 | 185 | YNRRLKKYALVDFGLAQ | EECCCCEEEEEECCCCC | c1c-AT-4 | uniq | 0.019 | 0.043 | 0.143 | boundary | MID|STKc_Cdc7; | 0.75 |
| 22 | fp_D | O00311 | 202 | GTHDTKIELLKFVQ | CCCCCCCHHCCCCC | c2-AT-4 | uniq | 0.383 | 0.47 | 0.25 | boundary | MID|STKc_Cdc7; | 0.0 |
| 23 | fp_D | O00311 | 351 | CDCYATDKVCSICLSR | CCCCCCCCCCCCCCCC | c1a-AT-4 | multi-selected | 0.64 | 0.417 | 0.099 | boundary | MID|STKc_Cdc7; MID|PKc_like superfamily; boundary|PKc_like superfamily; | 0.0 |
| 24 | fp_D | O00311 | 352 | DCYATDKVCSICLSR | CCCCCCCCCCCCCCC | c1b-AT-4 | multi | 0.639 | 0.415 | 0.098 | boundary | MID|STKc_Cdc7; MID|PKc_like superfamily; boundary|PKc_like superfamily; | 0.0 |
| 25 | fp_D | O00311 | 393 | TAIDMWSAGVIFLSL | CHHHHHHHHHHHHHH | c1b-AT-4 | multi | 0.01 | 0.024 | 0.106 | __ | MID|STKc_Cdc7; boundary|PKc_like superfamily; MID|PKc_like superfamily; | 0.0 |
| 26 | fp_O | O00311 | 396 | DMWSAGVIFLSLLS | HHHHHHHHHHHHHH | c2-AT-5 | multi-selected | 0.01 | 0.022 | 0.089 | ORD | MID|STKc_Cdc7; boundary|PKc_like superfamily; MID|PKc_like superfamily; | 0.0 |
| 27 | fp_O | O00311 | 396 | DMWSAGVIFLSLLS | HHHHHHHHHHHHHH | c3-AT | multi-selected | 0.01 | 0.022 | 0.089 | ORD | MID|STKc_Cdc7; boundary|PKc_like superfamily; MID|PKc_like superfamily; | 0.0 |
| 28 | fp_O | O00311 | 413 | PFYKASDDLTALAQ | CCCCCCCCHHHHHH | c3-AT | multi | 0.01 | 0.037 | 0.199 | ORD | boundary|STKc_Cdc7; boundary|PKc_like superfamily; MID|PKc_like superfamily; | 0.0 |
| 29 | fp_O | O00311 | 417 | ASDDLTALAQIMTIRG | CCCCHHHHHHHHHHHC | c1a-AT-4 | multi-selected | 0.01 | 0.033 | 0.28 | ORD | boundary|STKc_Cdc7; boundary|PKc_like superfamily; MID|PKc_like superfamily; | 0.0 |
| 30 | fp_O | O00311 | 417 | ASDDLTALAQIMTIRG | CCCCHHHHHHHHHHHC | c1d-4 | multi | 0.01 | 0.033 | 0.28 | ORD | boundary|STKc_Cdc7; boundary|PKc_like superfamily; MID|PKc_like superfamily; | 0.0 |
| 31 | cand_D | O00311 | 457 | DLRKLCERLRGMDS +++++++*++ | CHHHHHHHHCCCCC | c3-4 | uniq | 0.024 | 0.211 | 0.383 | boundary | boundary|STKc_Cdc7; MID|PKc_like superfamily; | 0.0 |
| 32 | cand_D | O00311 | 538 | VPDEAYDLLDKLLDLNP ++++**++++ | CCCHHHHHHHHHCCCCC | c1c-AT-4 | uniq | 0.015 | 0.111 | 0.369 | boundary | boundary|PKc_like superfamily; | 0.0 |
| 33 | fp_D | O00311 | 560 | AEEALLHPFFKDMSLXX | HHHHHCCCCCCCCCC | c1c-4-Ct | multi-selected | 0.061 | 0.228 | 0.286 | boundary | boundary|PKc_like superfamily; | 0.0 |
| 34 | fp_D | O00311 | 561 | EEALLHPFFKDMSLXX | HHHHCCCCCCCCCC | c1a-4-Ct | multi-selected | 0.062 | 0.231 | 0.283 | boundary | boundary|PKc_like superfamily; | 0.0 |
| 35 | fp_D | O00311 | 561 | EEALLHPFFKDMSLXX | HHHHCCCCCCCCCC | c1d-4-Ct | multi | 0.062 | 0.231 | 0.283 | boundary | boundary|PKc_like superfamily; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment