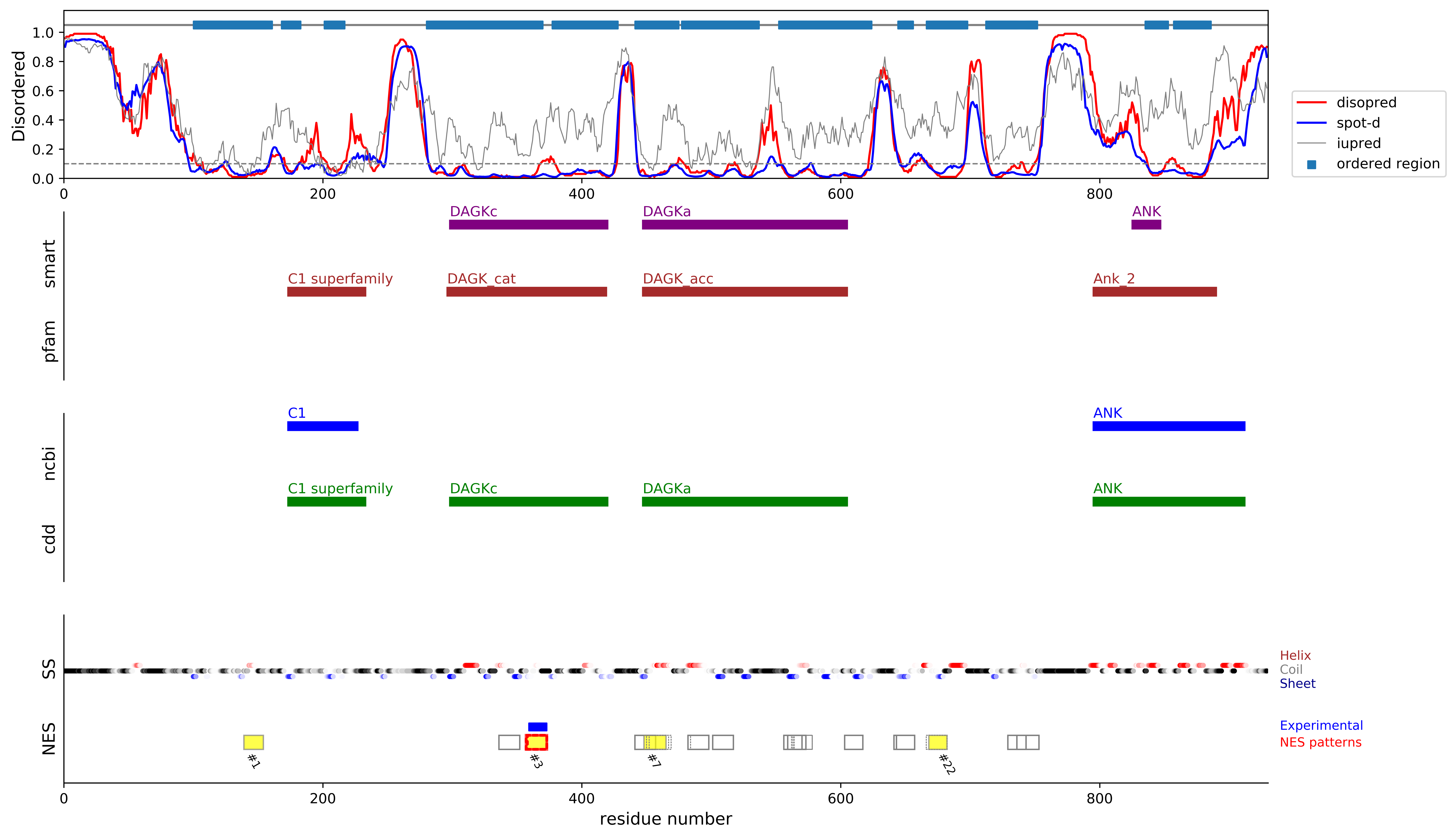

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | O08560 | 139 | HTPCIGQLEKINFRC | CCCCHHHHHHCCCCC | c1b-4 | uniq | 0.024 | 0.04 | 0.095 | boundary | boundary|C1 superfamily; | 0.0 |
| 2 | fp_O | O08560 | 336 | EALEMYRKVHNLRILA | HHHHHHHCCCCEEEEE | c1a-4 | uniq | 0.011 | 0.011 | 0.3 | ORD | MID|DAGKc; | 0.0 |
| 3 | cand_D | O08560 | 357 | TVGWILSTLDQLRLKP *++*++*+* | CEEEECCHHHHCCCCC | c1a-5 | multi-selected | 0.055 | 0.017 | 0.256 | boundary | MID|DAGKc; | 0.143 |
| 4 | cand_D | O08560 | 357 | TVGWILSTLDQLRLKP *++*++*+* | CEEEECCHHHHCCCCC | c1d-AT-5 | multi | 0.055 | 0.017 | 0.256 | boundary | MID|DAGKc; | 0.143 |
| 5 | cand_D | O08560 | 358 | VGWILSTLDQLRLKP *++*++*+* | EEEECCHHHHCCCCC | c1b-4 | multi | 0.058 | 0.017 | 0.262 | boundary | MID|DAGKc; | 0.0 |
| 6 | fp_beta_D | O08560 | 441 | DRLPLDVFNNYFSLGF | CCCCCEEEECCCCHHH | c1d-4 | uniq | 0.069 | 0.057 | 0.182 | boundary | boundary|DAGKc; boundary|DAGKa; | 0.571 |
| 7 | fp_D | O08560 | 448 | FNNYFSLGFDAHVTLEF | EECCCCHHHHHHHHHHH | c1c-4 | multi-selected | 0.027 | 0.021 | 0.169 | boundary | boundary|DAGKc; boundary|DAGKa; | 0.0 |
| 8 | fp_D | O08560 | 450 | NYFSLGFDAHVTLEFHE | CCCCHHHHHHHHHHHHH | c1c-AT-4 | multi | 0.028 | 0.023 | 0.2 | __ | boundary|DAGKc; boundary|DAGKa; | 0.0 |
| 9 | fp_D | O08560 | 452 | FSLGFDAHVTLEFHESR | CCHHHHHHHHHHHHHHH | c1cR-4 | multi | 0.033 | 0.029 | 0.245 | boundary | boundary|DAGKc; boundary|DAGKa; | 0.0 |
| 10 | fp_D | O08560 | 482 | KMFYAGTAFSDFLMGS | HHHHHHHHHHHHHHCC | c1a-AT-5 | multi-selected | 0.048 | 0.018 | 0.15 | __ | MID|DAGKa; | 0.0 |
| 11 | fp_D | O08560 | 484 | FYAGTAFSDFLMGS | HHHHHHHHHHHHCC | c2-AT-4 | multi | 0.044 | 0.018 | 0.144 | __ | MID|DAGKa; | 0.0 |
| 12 | fp_beta_O | O08560 | 501 | LAKHIRVVCDGMDLTP | CCCCEEEEECCCCCCC | c1d-4 | uniq | 0.041 | 0.034 | 0.244 | ORD | MID|DAGKa; | 0.714 |
| 13 | fp_beta_D | O08560 | 556 | DDGYLEVIGFTMTS | CCCEEEEEEEEHHH | c2-4 | multi-selected | 0.027 | 0.035 | 0.243 | boundary | MID|DAGKa; | 1.0 |
| 14 | fp_beta_D | O08560 | 559 | YLEVIGFTMTSLAA | EEEEEEEEHHHHHH | c3-4 | multi-selected | 0.028 | 0.034 | 0.218 | boundary | MID|DAGKa; | 0.714 |
| 15 | fp_D | O08560 | 562 | VIGFTMTSLAALQVGG | EEEEEHHHHHHHHCCC | c1a-AT-5 | multi | 0.041 | 0.052 | 0.236 | boundary | MID|DAGKa; | 0.143 |
| 16 | fp_D | O08560 | 563 | IGFTMTSLAALQVGG | EEEEHHHHHHHHCCC | c1b-4 | multi | 0.043 | 0.054 | 0.231 | boundary | MID|DAGKa; | 0.0 |
| 17 | fp_D | O08560 | 564 | GFTMTSLAALQVGG | EEEHHHHHHHHCCC | c2-AT-5 | multi | 0.045 | 0.057 | 0.234 | boundary | MID|DAGKa; | 0.0 |
| 18 | fp_D | O08560 | 603 | EPCKLAASRIRIAL | CCCCCCCCEEEEEE | c2-AT-4 | uniq | 0.012 | 0.023 | 0.298 | boundary | boundary|DAGKa; | 0.286 |
| 19 | fp_beta_D | O08560 | 641 | VPEQLRIQVSRVSMHD | CCCEEEEEEEEECCCC | c1a-4 | multi-selected | 0.105 | 0.135 | 0.429 | boundary | 1.0 | |
| 20 | fp_beta_D | O08560 | 643 | EQLRIQVSRVSMHD | CEEEEEEEEECCCC | c2-4 | multi-selected | 0.089 | 0.114 | 0.411 | boundary | 1.0 | |
| 21 | fp_D | O08560 | 666 | QLKEASVPLGTVVVPG | HHHHCCCCCEEEEECC | c1a-AT-5 | multi | 0.048 | 0.056 | 0.392 | boundary | 0.286 | |
| 22 | fp_D | O08560 | 668 | KEASVPLGTVVVPG | HHCCCCCEEEEECC | c2-4 | multi-selected | 0.039 | 0.049 | 0.397 | boundary | 0.429 | |
| 23 | fp_D | O08560 | 729 | RIDRAQEHLNYVTE | CCCCCCCCCCCHHH | c3-AT | uniq | 0.079 | 0.033 | 0.323 | boundary | 0.0 | |
| 24 | fp_D | O08560 | 736 | HLNYVTEIAQDEIYILD | CCCCHHHCCCCCCEECC | c1c-AT-5 | uniq | 0.081 | 0.027 | 0.291 | boundary | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment