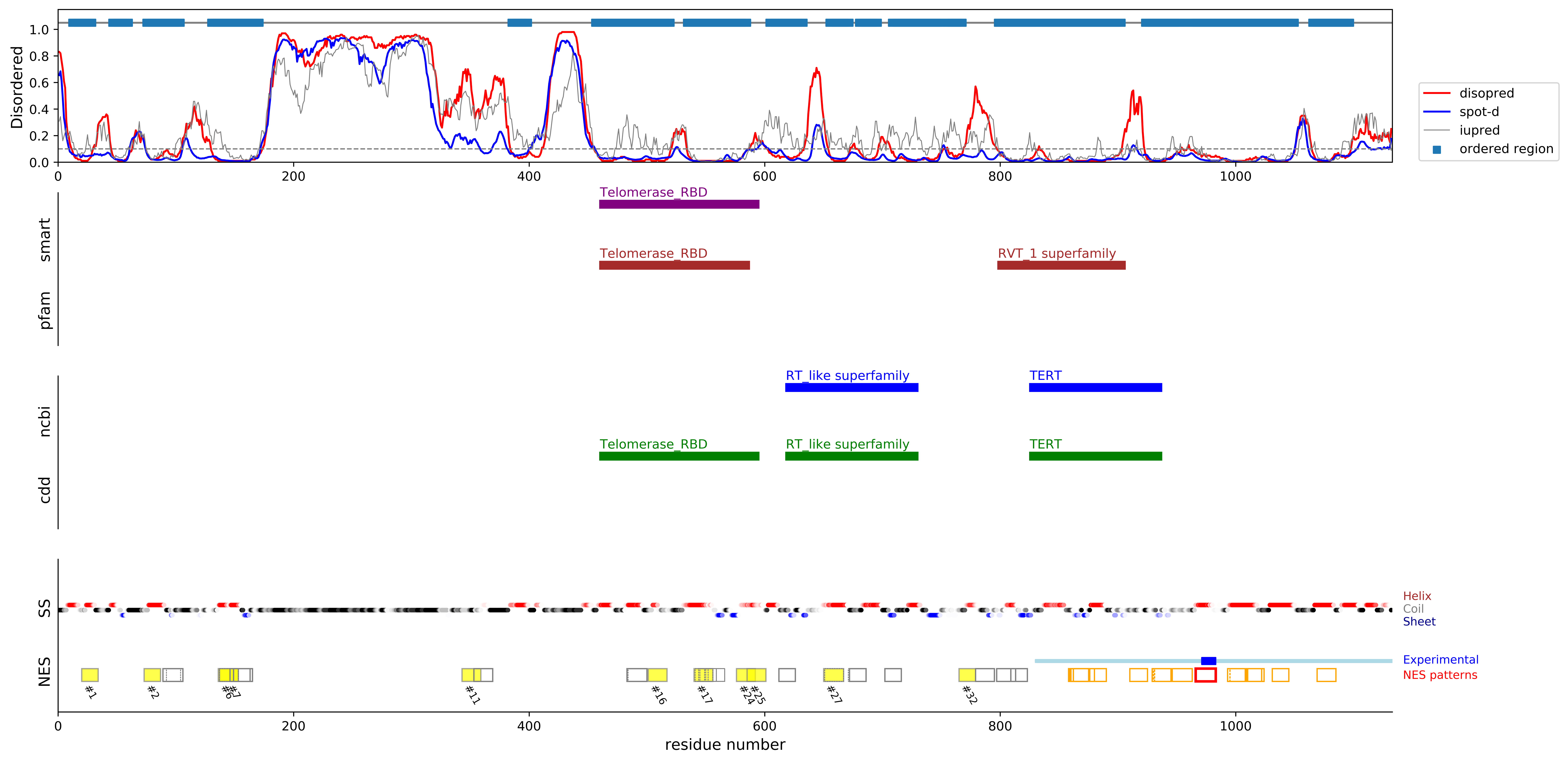

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | O14746 | 20 | EVLPLATFVRRLGP | CCCCHHHHHHHHCC | c3-4 | uniq | 0.055 | 0.05 | 0.117 | boundary | 0.0 | |
| 2 | fp_D | O14746 | 73 | QVSCLKELVARVLQ | CCCCHHHHHHHHHH | c3-4 | uniq | 0.05 | 0.043 | 0.067 | boundary | 0.0 | |
| 3 | fp_D | O14746 | 89 | CERGAKNVLAFGFALLD | HHCCCCCEECCCCCCCC | c1c-AT-4 | multi-selected | 0.068 | 0.027 | 0.079 | boundary | 0.25 | |
| 4 | fp_D | O14746 | 89 | CERGAKNVLAFGFAL | HHCCCCCEECCCCCC | c1b-AT-4 | multi | 0.063 | 0.023 | 0.071 | boundary | 0.286 | |
| 5 | fp_D | O14746 | 92 | GAKNVLAFGFALLD | CCCCEECCCCCCCC | c2-AT-4 | multi | 0.068 | 0.027 | 0.088 | boundary | 0.286 | |
| 6 | fp_D | O14746 | 136 | AWGLLLRRVGDDVLVHL | HHHHHHHHHHHHHHHHH | c1c-4 | multi-selected | 0.01 | 0.013 | 0.099 | boundary | 0.0 | |
| 7 | fp_D | O14746 | 137 | WGLLLRRVGDDVLVHL | HHHHHHHHHHHHHHHH | c1aR-4 | multi-selected | 0.01 | 0.012 | 0.091 | boundary | 0.0 | |
| 8 | fp_D | O14746 | 137 | WGLLLRRVGDDVLVHL | HHHHHHHHHHHHHHHH | c1d-4 | multi | 0.01 | 0.012 | 0.091 | boundary | 0.0 | |
| 9 | fp_D | O14746 | 146 | DDVLVHLLARCALFVLV | HHHHHHHHHHCCEEEEE | c1c-AT-4 | multi-selected | 0.01 | 0.008 | 0.03 | boundary | 0.0 | |
| 10 | fp_D | O14746 | 149 | LVHLLARCALFVLVAP | HHHHHHHCCEEEEECC | c1a-AT-5 | multi-selected | 0.011 | 0.009 | 0.029 | boundary | 0.286 | |
| 11 | fp_D | O14746 | 343 | PSFLLSSLRPSLTGAR | CCCCCCCCCCCCCCCC | c1aR-4 | uniq | 0.532 | 0.157 | 0.417 | DISO | 0.0 | |
| 12 | fp_D | O14746 | 353 | SLTGARRLVETIFLGS | CCCCCCCHHHHHHCCC | c1a-AT-5 | multi-selected | 0.429 | 0.102 | 0.356 | DISO | 0.0 | |
| 13 | fp_D | O14746 | 353 | SLTGARRLVETIFLGS | CCCCCCCHHHHHHCCC | c1d-AT-5 | multi | 0.429 | 0.102 | 0.356 | DISO | 0.0 | |
| 14 | fp_O | O14746 | 483 | NERRFLRNTKKFISLGK | HHHHHHHHHHHHHHCCC | c1c-AT-4 | multi-selected | 0.014 | 0.026 | 0.201 | ORD | MID|Telomerase_RBD; | 0.0 |
| 15 | fp_O | O14746 | 484 | ERRFLRNTKKFISLGK | HHHHHHHHHHHHHCCC | c1d-AT-4 | multi | 0.013 | 0.026 | 0.192 | ORD | MID|Telomerase_RBD; | 0.0 |
| 16 | fp_D | O14746 | 501 | AKLSLQELTWKMSVRD | CCCCHHHHHCCCCCCC | c1aR-4 | uniq | 0.013 | 0.03 | 0.105 | boundary | MID|Telomerase_RBD; | 0.0 |
| 17 | fp_D | O14746 | 540 | ILAKFLHWLMSVYVVE | HHHHHHHHHHHHHHHH | c1a-5 | multi-selected | 0.01 | 0.007 | 0.014 | boundary | MID|Telomerase_RBD; | 0.0 |
| 18 | fp_D | O14746 | 541 | LAKFLHWLMSVYVVE | HHHHHHHHHHHHHHH | c1b-4 | multi | 0.01 | 0.006 | 0.011 | boundary | MID|Telomerase_RBD; | 0.0 |
| 19 | fp_D | O14746 | 544 | FLHWLMSVYVVELLR | HHHHHHHHHHHHCCC | c1b-5 | multi | 0.01 | 0.006 | 0.008 | boundary | MID|Telomerase_RBD; | 0.0 |
| 20 | fp_D | O14746 | 545 | LHWLMSVYVVELLR | HHHHHHHHHHHCCC | c2-4 | multi | 0.01 | 0.006 | 0.008 | boundary | MID|Telomerase_RBD; | 0.0 |
| 21 | fp_O | O14746 | 549 | MSVYVVELLRSFFYVTE | HHHHHHHCCCEEEEEEC | c1c-4 | multi | 0.01 | 0.007 | 0.013 | ORD | MID|Telomerase_RBD; | 0.25 |
| 22 | fp_O | O14746 | 550 | SVYVVELLRSFFYVTE | HHHHHHCCCEEEEEEC | c1d-5 | multi | 0.01 | 0.007 | 0.013 | ORD | MID|Telomerase_RBD; | 0.286 |
| 23 | fp_O | O14746 | 552 | YVVELLRSFFYVTE | HHHHCCCEEEEEEC | c3-4 | multi | 0.01 | 0.007 | 0.014 | ORD | MID|Telomerase_RBD; | 0.429 |
| 24 | fp_D | O14746 | 576 | YRKSVWSKLQSIGIRQ | EEEHHHHHHHHHHHHH | c1a-4 | uniq | 0.076 | 0.061 | 0.058 | boundary | boundary|Telomerase_RBD; | 0.0 |
| 25 | fp_D | O14746 | 585 | QSIGIRQHLKRVQLRE | HHHHHHHHHHHHHHHC | c1a-4 | uniq | 0.144 | 0.108 | 0.141 | boundary | boundary|Telomerase_RBD; boundary|RT_like superfamily; | 0.0 |
| 26 | fp_D | O14746 | 612 | ARPALLTSRLRFIP | CCCCCCCCCEEEEE | c2-AT-4 | uniq | 0.037 | 0.045 | 0.214 | boundary | boundary|Telomerase_RBD; boundary|RT_like superfamily; | 0.143 |
| 27 | fp_D | O14746 | 650 | RAERLTSRVKALFSVLN | HHHHHHHHHHHHHHHHH | c1c-4 | multi-selected | 0.072 | 0.046 | 0.184 | boundary | MID|RT_like superfamily; | 0.0 |
| 28 | fp_D | O14746 | 651 | AERLTSRVKALFSVLN | HHHHHHHHHHHHHHHH | c1d-AT-4 | multi | 0.056 | 0.04 | 0.174 | boundary | MID|RT_like superfamily; | 0.0 |
| 29 | fp_D | O14746 | 671 | RRPGLLGASVLGLDD | CCCCCCCCCCCCHHH | c1b-AT-4 | multi | 0.084 | 0.052 | 0.147 | boundary | MID|RT_like superfamily; | 0.0 |
| 30 | fp_D | O14746 | 672 | RPGLLGASVLGLDD | CCCCCCCCCCCHHH | c2-AT-4 | multi-selected | 0.086 | 0.052 | 0.153 | boundary | MID|RT_like superfamily; | 0.0 |
| 31 | fp_beta_D | O14746 | 702 | PPPELYFVKVDVTG | CCCCEEEEECCCCC | c2-4 | uniq | 0.085 | 0.025 | 0.198 | __ | boundary|RT_like superfamily; | 0.714 |
| 32 | fp_D | O14746 | 765 | TLTDLQPYMRQFVA | CCCCCCHHHHHHHH | c3-4 | uniq | 0.206 | 0.044 | 0.162 | boundary | 0.0 | |
| 33 | fp_D | O14746 | 779 | HLQETSPLRDAVVIEQ | HHHCCCCCCCCEEEEC | c1d-AT-5 | uniq | 0.366 | 0.049 | 0.226 | boundary | 0.0 | |
| 34 | fp_D | O14746 | 797 | SLNEASSGLFDVFLRF | CEEECCCCHHHHHHHH | c1a-AT-5 | uniq | 0.039 | 0.037 | 0.053 | boundary | boundary|TERT; | 0.0 |
| 35 | fp_D | O14746 | 809 | FLRFMCHHAVRIRG | HHHHHHCCEEEECC | c3-AT | uniq | 0.011 | 0.01 | 0.027 | boundary | boundary|TERT; | 0.286 |
| 36 | fp_beta_O | O14746 | 858 | RRDGLLLRLVDDFLLVT ................. | CCCCCEEEEECCEEEEE | c1c-4 | multi-selected | 0.011 | 0.012 | 0.031 | ORD | MID|TERT; | 0.625 |
| 37 | fp_beta_O | O14746 | 859 | RDGLLLRLVDDFLLVT ................ | CCCCEEEEECCEEEEE | c1a-4 | multi-selected | 0.01 | 0.011 | 0.029 | ORD | MID|TERT; | 0.714 |
| 38 | fp_beta_O | O14746 | 859 | RDGLLLRLVDDFLLVT ................ | CCCCEEEEECCEEEEE | c1d-4 | multi | 0.01 | 0.011 | 0.029 | ORD | MID|TERT; | 0.714 |
| 39 | fp_beta_O | O14746 | 859 | RDGLLLRLVDDFLLVTP ................. | CCCCEEEEECCEEEEEC | c1c-4 | multi-selected | 0.01 | 0.011 | 0.03 | ORD | MID|TERT; | 0.75 |
| 40 | fp_beta_O | O14746 | 860 | DGLLLRLVDDFLLVT ............... | CCCEEEEECCEEEEE | c1b-4 | multi | 0.01 | 0.009 | 0.027 | ORD | MID|TERT; | 0.714 |
| 41 | fp_beta_O | O14746 | 860 | DGLLLRLVDDFLLVTP ................ | CCCEEEEECCEEEEEC | c1d-4 | multi | 0.01 | 0.009 | 0.028 | ORD | MID|TERT; | 0.714 |
| 42 | fp_beta_O | O14746 | 860 | DGLLLRLVDDFLLVTP ................ | CCCEEEEECCEEEEEC | c1aR-4 | multi-selected | 0.01 | 0.009 | 0.028 | ORD | MID|TERT; | 0.714 |
| 43 | fp_beta_O | O14746 | 862 | LLLRLVDDFLLVTPHLTH .................. | CEEEEECCEEEEECCHHH | c4-5 | multi-selected | 0.01 | 0.009 | 0.032 | ORD | MID|TERT; | 0.75 |
| 44 | fp_beta_O | O14746 | 862 | LLLRLVDDFLLVTP .............. | CEEEEECCEEEEEC | c3-4 | multi-selected | 0.01 | 0.006 | 0.028 | ORD | MID|TERT; | 0.714 |
| 45 | fp_D | O14746 | 876 | HLTHAKTFLRTLVR .............. | CHHHHHHHHHHHHH | c3-AT | uniq | 0.01 | 0.017 | 0.08 | boundary | MID|TERT; | 0.0 |
| 46 | fp_D | O14746 | 910 | EDEALGGTAFVQMPA ............... | CCCCCCCCCCCCCCC | c1b-AT-4 | uniq | 0.35 | 0.081 | 0.077 | boundary | boundary|TERT; | 0.0 |
| 47 | fp_beta_D | O14746 | 929 | PWCGLLLDTRTLEVQS ................ | CCCCEEEECCCCEEEC | c1a-AT-4 | multi-selected | 0.014 | 0.017 | 0.037 | boundary | boundary|TERT; | 0.571 |
| 48 | fp_D | O14746 | 930 | WCGLLLDTRTLEVQS ............... | CCCEEEECCCCEEEC | c1b-AT-4 | multi | 0.014 | 0.018 | 0.038 | boundary | boundary|TERT; | 0.429 |
| 49 | fp_D | O14746 | 931 | CGLLLDTRTLEVQS .............. | CCEEEECCCCEEEC | c2-AT-4 | multi | 0.014 | 0.018 | 0.04 | __ | boundary|TERT; | 0.429 |
| 50 | fp_O | O14746 | 946 | YSSYARTSIRASLTFNR ................. | CCCCCCCCCCCCCCCCC | c1c-AT-4 | uniq | 0.084 | 0.053 | 0.118 | ORD | boundary|TERT; | 0.0 |
| 51 | cand_O | O14746 | 966 | AGRNMRRKLFGVLRLKC ........*...*.*.. | HHHHHHHHHHHHHHHCC | c1c-4 | uniq | 0.051 | 0.019 | 0.051 | ORD | boundary|TERT; | 0.0 |
| 52 | fp_O | O14746 | 993 | SLQTVCTNIYKILLLQA ................. | CHHHHHHHHHHHHHHHH | c1c-5 | multi | 0.011 | 0.007 | 0.011 | ORD | 0.0 | |
| 53 | fp_O | O14746 | 993 | SLQTVCTNIYKILLLQ ................ | CHHHHHHHHHHHHHHH | c1a-5 | multi-selected | 0.01 | 0.007 | 0.011 | ORD | 0.0 | |
| 54 | fp_O | O14746 | 995 | QTVCTNIYKILLLQ .............. | HHHHHHHHHHHHHH | c2-AT-4 | multi | 0.01 | 0.005 | 0.01 | ORD | 0.0 | |
| 55 | fp_O | O14746 | 1008 | QAYRFHACVLQLPF .............. | HHHHHHHHHHHCCC | c2-AT-4 | multi-selected | 0.026 | 0.01 | 0.015 | ORD | 0.0 | |
| 56 | fp_O | O14746 | 1010 | YRFHACVLQLPFHQ .............. | HHHHHHHHHCCCCC | c2-AT-4 | multi-selected | 0.029 | 0.016 | 0.016 | ORD | 0.0 | |
| 57 | fp_D | O14746 | 1031 | FFLRVISDTASLCY .............. | HHHHHHHHHHHHHH | c3-AT | uniq | 0.01 | 0.013 | 0.026 | boundary | 0.0 | |
| 58 | fp_D | O14746 | 1069 | AVQWLCHQAFLLKLTR ................ | HHHHHHHHHHHHHHHC | c1a-AT-5 | uniq | 0.017 | 0.016 | 0.025 | boundary | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment