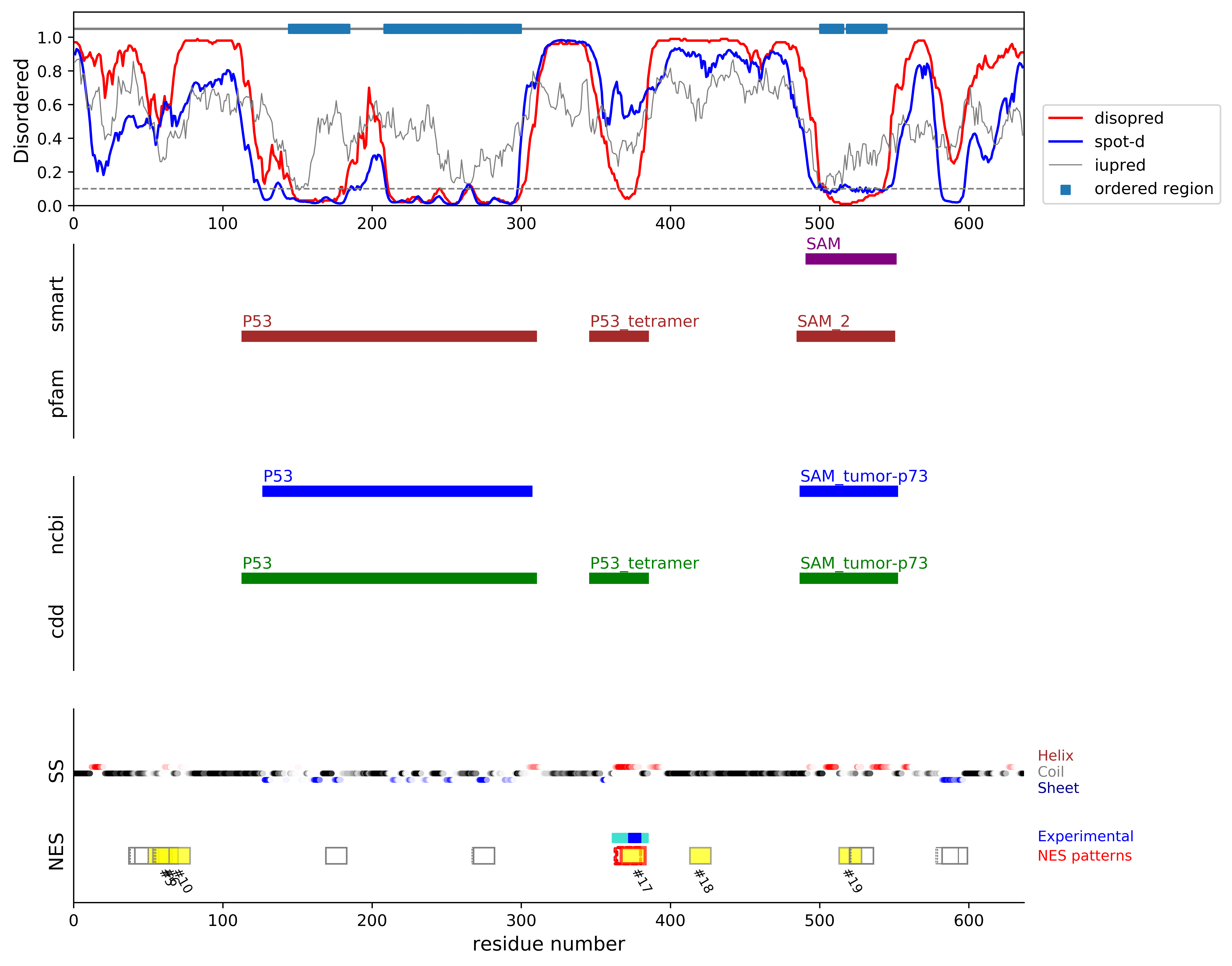

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | O15350 | 37 | GNNEVVGGTDSSMDVFH | CCCCCCCCCCCCCCCCC | c1c-AT-4 | multi-selected | 0.794 | 0.495 | 0.63 | DISO | 0.0 | |
| 2 | fp_D | O15350 | 38 | NNEVVGGTDSSMDVFH | CCCCCCCCCCCCCCCC | c1d-AT-4 | multi | 0.784 | 0.492 | 0.627 | DISO | 0.0 | |
| 3 | fp_D | O15350 | 41 | VVGGTDSSMDVFHLEG | CCCCCCCCCCCCCCCC | c1a-AT-5 | multi-selected | 0.719 | 0.468 | 0.565 | DISO | 0.0 | |
| 4 | fp_D | O15350 | 50 | DVFHLEGMTTSVMA | CCCCCCCCCCHHHH | c3-AT | multi | 0.578 | 0.501 | 0.378 | DISO | 0.0 | |
| 5 | fp_D | O15350 | 50 | DVFHLEGMTTSVMAQF | CCCCCCCCCCHHHHHH | c1aR-4 | multi-selected | 0.58 | 0.512 | 0.384 | DISO | 0.0 | |
| 6 | fp_D | O15350 | 53 | HLEGMTTSVMAQFNLLS | CCCCCCCHHHHHHCCCC | c1c-5 | multi-selected | 0.626 | 0.512 | 0.375 | DISO | 0.0 | |
| 7 | fp_D | O15350 | 54 | LEGMTTSVMAQFNLLS | CCCCCCHHHHHHCCCC | c1a-AT-4 | multi | 0.626 | 0.514 | 0.368 | DISO | 0.0 | |
| 8 | fp_D | O15350 | 54 | LEGMTTSVMAQFNLLS | CCCCCCHHHHHHCCCC | c1d-AT-4 | multi | 0.626 | 0.514 | 0.368 | DISO | 0.0 | |
| 9 | fp_D | O15350 | 55 | EGMTTSVMAQFNLLS | CCCCCHHHHHHCCCC | c1b-AT-4 | multi | 0.623 | 0.52 | 0.359 | DISO | 0.0 | |
| 10 | fp_D | O15350 | 64 | QFNLLSSTMDQMSS | HHCCCCCHHHHHCC | c3-4 | uniq | 0.851 | 0.553 | 0.439 | DISO | 0.0 | |
| 11 | fp_beta_D | O15350 | 169 | PPPGTAIRAMPVYK | CCCCCEEEEEECCC | c2-AT-4 | uniq | 0.053 | 0.027 | 0.498 | boundary | MID|P53; | 0.714 |
| 12 | fp_beta_D | O15350 | 267 | NRRPILIIITLEMRD | CCCCEEEEEEECCCC | c1b-4 | multi | 0.039 | 0.027 | 0.244 | boundary | MID|P53; | 1.0 |
| 13 | fp_beta_D | O15350 | 268 | RRPILIIITLEMRD | CCCEEEEEEECCCC | c2-4 | multi-selected | 0.034 | 0.022 | 0.251 | boundary | MID|P53; | 1.0 |
| 14 | cand_D | O15350 | 363 | ENFEILMKLKESLELME +++++++++++*+*++ | HHHHHHHHHHHHHHHHH | c1c-4 | multi | 0.089 | 0.59 | 0.35 | DISO | small|P53_tetramer; | 0.0 |
| 15 | fp_D | O15350 | 363 | ENFEILMKLKESLELME +++++++++++*+*++ | HHHHHHHHHHHHHHHHH | c1cR-5 | multi | 0.089 | 0.59 | 0.35 | DISO | small|P53_tetramer; | 0.0 |
| 16 | cand_D | O15350 | 364 | NFEILMKLKESLELME +++++++++++*+*++ | HHHHHHHHHHHHHHHH | c1d-5 | multi | 0.086 | 0.583 | 0.352 | DISO | small|P53_tetramer; | 0.0 |
| 17 | cand_D | O15350 | 367 | ILMKLKESLELMELVP ++++++++*+*+++++ | HHHHHHHHHHHHHCCC | c1a-5 | multi-selected | 0.12 | 0.569 | 0.39 | DISO | small|P53_tetramer; | 0.0 |
| 18 | fp_D | O15350 | 413 | PMNKVHGGMNKLPS | CCCCCCCCCCCCCC | c3-4 | uniq | 0.979 | 0.86 | 0.607 | DISO | 0.0 | |
| 19 | fp_D | O15350 | 513 | GLQSIYHLQNLTIED | CCCCCCCCCCCCHHH | c1b-5 | uniq | 0.016 | 0.103 | 0.254 | boundary | MID|SAM_tumor-p73; | 0.0 |
| 20 | fp_D | O15350 | 520 | LQNLTIEDLGALKIPE | CCCCCHHHHCCCCCCH | c1a-AT-4 | multi-selected | 0.034 | 0.092 | 0.297 | boundary | boundary|SAM_tumor-p73; | 0.0 |
| 21 | fp_D | O15350 | 521 | QNLTIEDLGALKIPE | CCCCHHHHCCCCCCH | c1b-4 | multi | 0.035 | 0.091 | 0.292 | boundary | boundary|SAM_tumor-p73; | 0.0 |
| 22 | fp_beta_D | O15350 | 578 | QRQRVMEAVHFRVRH | CCCCEEEEEEEEEEE | c1b-AT-4 | multi | 0.451 | 0.088 | 0.351 | DISO | boundary|SAM_tumor-p73; | 1.0 |
| 23 | fp_beta_D | O15350 | 579 | RQRVMEAVHFRVRH | CCCEEEEEEEEEEE | c2-AT-4 | multi | 0.434 | 0.063 | 0.352 | DISO | boundary|SAM_tumor-p73; | 1.0 |
| 24 | fp_beta_D | O15350 | 582 | VMEAVHFRVRHTITIPN | EEEEEEEEEEEEEEECC | c1c-5 | multi-selected | 0.391 | 0.044 | 0.413 | DISO | boundary|SAM_tumor-p73; | 1.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment