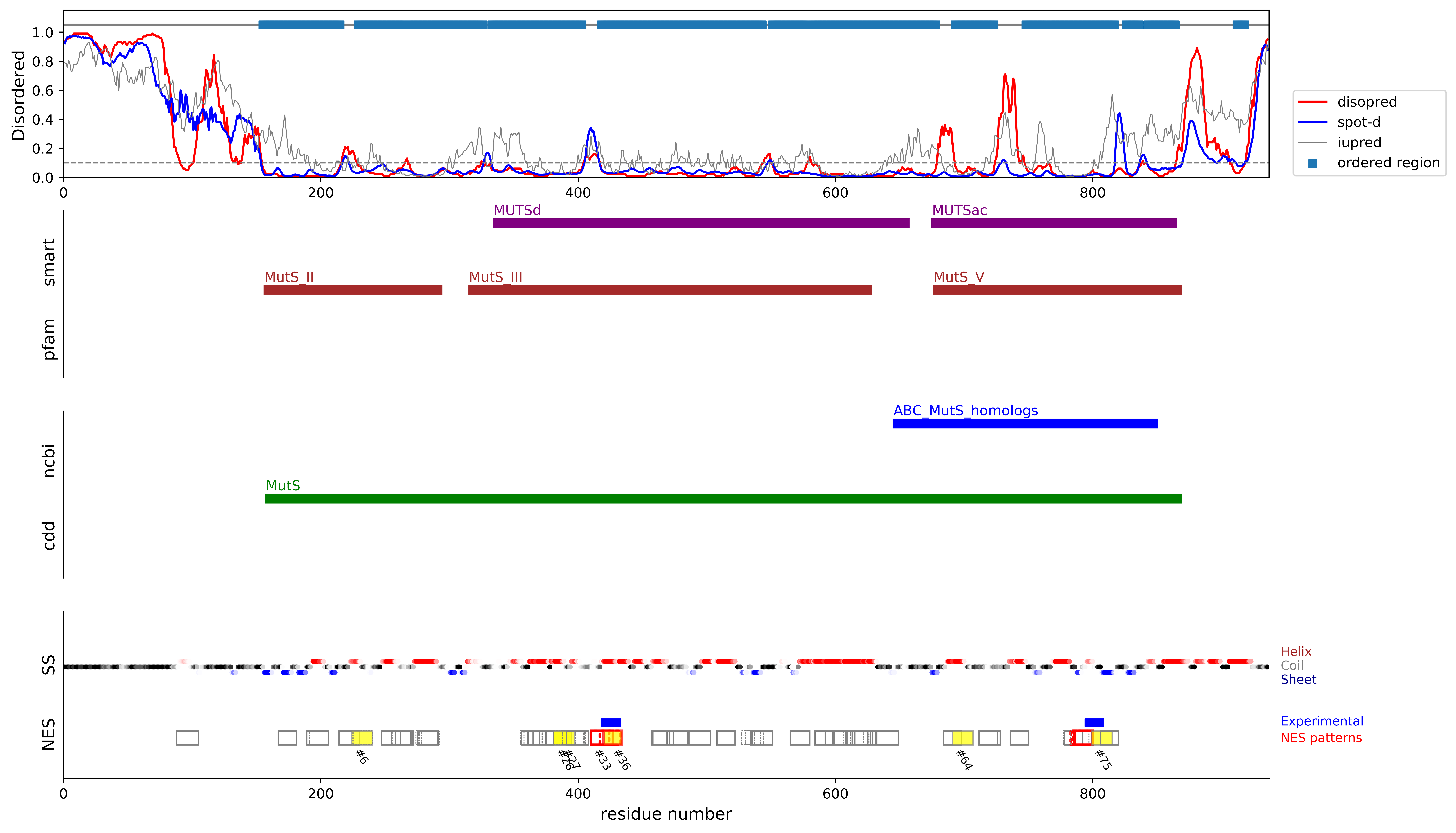

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | O15457 | 88 | KRAYAENTVASNFTFGA | CCHHHHHHHHHCCCCCC | c1c-AT-4 | uniq | 0.114 | 0.447 | 0.443 | DISO | 0.0 | |
| 2 | fp_beta_D | O15457 | 167 | ARGEIGMASIDLKN | CCCCEEEEEEECCC | c2-4 | uniq | 0.01 | 0.021 | 0.232 | boundary | boundary|MutS; | 0.857 |
| 3 | fp_D | O15457 | 189 | ADNTTYAKVITKLKILS | CCCCHHHHHHHHHHCCC | c1c-AT-4 | multi-selected | 0.01 | 0.027 | 0.075 | boundary | MID|MutS; | 0.0 |
| 4 | fp_D | O15457 | 191 | NTTYAKVITKLKILS | CCHHHHHHHHHHCCC | c1b-AT-4 | multi | 0.01 | 0.026 | 0.066 | boundary | MID|MutS; | 0.0 |
| 5 | fp_D | O15457 | 214 | TACAVGNSTKLFTLIT | CCCCCCCCHHHHHHHH | c1a-AT-4 | uniq | 0.146 | 0.076 | 0.088 | boundary | MID|MutS; | 0.0 |
| 6 | fp_D | O15457 | 224 | LFTLITENFKNVNFTT | HHHHHHHHCCCCCCEE | c1a-5 | multi-selected | 0.082 | 0.039 | 0.094 | boundary | MID|MutS; | 0.0 |
| 7 | fp_D | O15457 | 225 | FTLITENFKNVNFTT | HHHHHHHCCCCCCEE | c1b-AT-4 | multi | 0.075 | 0.039 | 0.094 | boundary | MID|MutS; | 0.0 |
| 8 | fp_O | O15457 | 247 | ETKGLEYIEQLCIAE | HHHHHHHHHHHHHHC | c1b-4 | uniq | 0.024 | 0.055 | 0.049 | ORD | MID|MutS; | 0.0 |
| 9 | fp_O | O15457 | 255 | EQLCIAEFSTVLMEV | HHHHHHCCCCCCCCC | c1b-4 | multi | 0.064 | 0.046 | 0.027 | ORD | MID|MutS; | 0.0 |
| 10 | fp_O | O15457 | 255 | EQLCIAEFSTVLMEVQ | HHHHHHCCCCCCCCCC | c1aR-5 | multi-selected | 0.063 | 0.045 | 0.026 | ORD | MID|MutS; | 0.0 |
| 11 | fp_O | O15457 | 256 | QLCIAEFSTVLMEV | HHHHHCCCCCCCCC | c2-AT-5 | multi | 0.067 | 0.046 | 0.026 | ORD | MID|MutS; | 0.0 |
| 12 | fp_O | O15457 | 258 | CIAEFSTVLMEVQS | HHHCCCCCCCCCCC | c2-AT-5 | multi | 0.071 | 0.046 | 0.022 | ORD | MID|MutS; | 0.0 |
| 13 | fp_O | O15457 | 258 | CIAEFSTVLMEVQS | HHHCCCCCCCCCCC | c3-4 | multi-selected | 0.071 | 0.046 | 0.022 | ORD | MID|MutS; | 0.0 |
| 14 | fp_O | O15457 | 274 | YCLAAVAALLKYVEFIQ | HHHHHHHHHHHHHHHHH | c1c-AT-4 | multi | 0.012 | 0.015 | 0.013 | ORD | MID|MutS; | 0.0 |
| 15 | fp_O | O15457 | 275 | CLAAVAALLKYVEFIQ | HHHHHHHHHHHHHHHH | c1a-5 | multi-selected | 0.011 | 0.015 | 0.013 | ORD | MID|MutS; | 0.0 |
| 16 | fp_O | O15457 | 275 | CLAAVAALLKYVEFIQ | HHHHHHHHHHHHHHHH | c1d-5 | multi | 0.011 | 0.015 | 0.013 | ORD | MID|MutS; | 0.0 |
| 17 | fp_O | O15457 | 276 | LAAVAALLKYVEFIQ | HHHHHHHHHHHHHHH | c1b-AT-4 | multi | 0.011 | 0.015 | 0.013 | ORD | MID|MutS; | 0.0 |
| 18 | fp_O | O15457 | 277 | AAVAALLKYVEFIQ | HHHHHHHHHHHHHH | c2-AT-4 | multi | 0.011 | 0.015 | 0.013 | ORD | MID|MutS; | 0.0 |
| 19 | fp_O | O15457 | 278 | AVAALLKYVEFIQN | HHHHHHHHHHHHHC | c3-4 | multi | 0.013 | 0.017 | 0.013 | ORD | MID|MutS; | 0.0 |
| 20 | fp_O | O15457 | 355 | ILEPLVDIETINMRL | HHCCCCCHHHHHHHH | c1b-5 | multi | 0.025 | 0.032 | 0.103 | ORD | MID|MutS; | 0.0 |
| 21 | fp_O | O15457 | 356 | LEPLVDIETINMRLDC | HCCCCCHHHHHHHHHH | c1a-AT-4 | multi | 0.021 | 0.03 | 0.1 | ORD | MID|MutS; | 0.0 |
| 22 | fp_O | O15457 | 356 | LEPLVDIETINMRL | HCCCCCHHHHHHHH | c2-4 | multi-selected | 0.022 | 0.032 | 0.095 | ORD | MID|MutS; | 0.0 |
| 23 | fp_O | O15457 | 358 | PLVDIETINMRLDC | CCCCHHHHHHHHHH | c2-AT-5 | multi | 0.017 | 0.03 | 0.089 | ORD | MID|MutS; | 0.0 |
| 24 | fp_O | O15457 | 361 | DIETINMRLDCVQE | CHHHHHHHHHHHHH | c3-4 | multi-selected | 0.014 | 0.026 | 0.075 | ORD | MID|MutS; | 0.0 |
| 25 | fp_O | O15457 | 365 | INMRLDCVQELLQDEE | HHHHHHHHHHHHHCHH | c1aR-4 | multi-selected | 0.013 | 0.023 | 0.062 | ORD | MID|MutS; | 0.0 |
| 26 | fp_D | O15457 | 381 | LFFGLQSVISRFLD | HHHHHHHHHCCCCC | c3-4 | multi-selected | 0.022 | 0.024 | 0.039 | boundary | MID|MutS; | 0.0 |
| 27 | fp_D | O15457 | 381 | LFFGLQSVISRFLDTE | HHHHHHHHHCCCCCHH | c1aR-4 | multi-selected | 0.023 | 0.024 | 0.039 | boundary | MID|MutS; | 0.0 |
| 28 | fp_D | O15457 | 381 | LFFGLQSVISRFLDTEQ | HHHHHHHHHCCCCCHHH | c1cR-4 | multi | 0.023 | 0.024 | 0.04 | boundary | MID|MutS; | 0.0 |
| 29 | fp_D | O15457 | 388 | VISRFLDTEQLLSVLV | HHCCCCCHHHHHHHCC | c1d-AT-5 | multi | 0.032 | 0.027 | 0.066 | boundary | MID|MutS; | 0.0 |
| 30 | fp_D | O15457 | 391 | RFLDTEQLLSVLVQI | CCCCHHHHHHHCCCC | c1b-AT-5 | multi | 0.045 | 0.036 | 0.092 | boundary | MID|MutS; | 0.0 |
| 31 | fp_D | O15457 | 391 | RFLDTEQLLSVLVQIPK | CCCCHHHHHHHCCCCCC | c1c-AT-5 | multi-selected | 0.056 | 0.055 | 0.106 | boundary | MID|MutS; | 0.0 |
| 32 | fp_D | O15457 | 391 | RFLDTEQLLSVLVQ | CCCCHHHHHHHCCC | c3-AT | multi | 0.039 | 0.031 | 0.083 | boundary | MID|MutS; | 0.0 |
| 33 | cand_D | O15457 | 410 | TVNAAESKITNLIYLKH ** * | CCHHCCCCHHHHHHHHH | c1c-AT-5 | multi-selected | 0.083 | 0.127 | 0.147 | boundary | MID|MutS; | 0.0 |
| 34 | cand_D | O15457 | 410 | TVNAAESKITNLIY ** | CCHHCCCCHHHHHH | c3-AT | multi | 0.096 | 0.148 | 0.164 | boundary | MID|MutS; | 0.0 |
| 35 | cand_D | O15457 | 417 | KITNLIYLKHTLELVD ** * * * | CHHHHHHHHHHHHHHH | c1d-5 | multi | 0.031 | 0.037 | 0.083 | boundary | MID|MutS; | 0.0 |
| 36 | cand_D | O15457 | 420 | NLIYLKHTLELVDP ** * * * | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.023 | 0.032 | 0.065 | boundary | MID|MutS; | 0.0 |
| 37 | fp_O | O15457 | 457 | RFGIILEKIKTVIN | CHHHHHHHHHHHHC | c3-4 | multi-selected | 0.033 | 0.036 | 0.062 | ORD | MID|MutS; | 0.0 |
| 38 | fp_O | O15457 | 458 | FGIILEKIKTVINDDA | HHHHHHHHHHHHCCCC | c1aR-4 | multi-selected | 0.031 | 0.038 | 0.071 | ORD | MID|MutS; | 0.0 |
| 39 | fp_O | O15457 | 469 | INDDARYMKGCLNMRT | HCCCCCCCCCCCCCCC | c1d-AT-4 | uniq | 0.023 | 0.053 | 0.127 | ORD | MID|MutS; | 0.0 |
| 40 | fp_O | O15457 | 486 | KCYAVRSNINEFLDIAR | EEEEECCCCCHHHHHHH | c1c-4 | uniq | 0.012 | 0.035 | 0.104 | ORD | MID|MutS; | 0.125 |
| 41 | fp_O | O15457 | 508 | IVDDIAGMISQLGE | HHHHHHHHHHHHHH | c3-4 | uniq | 0.029 | 0.03 | 0.108 | ORD | MID|MutS; | 0.0 |
| 42 | fp_beta_D | O15457 | 527 | LRTSFSSARGFFIQM | EEEEECCCEEEEEEE | c1b-AT-4 | multi | 0.02 | 0.025 | 0.055 | boundary | MID|MutS; | 0.571 |
| 43 | fp_beta_D | O15457 | 530 | SFSSARGFFIQMTT | EECCCEEEEEEEEC | c3-AT | multi | 0.029 | 0.023 | 0.052 | boundary | MID|MutS; | 0.714 |
| 44 | fp_beta_D | O15457 | 534 | ARGFFIQMTTDCIALPS | CEEEEEEEECCCCCCCC | c1c-AT-4 | multi-selected | 0.076 | 0.049 | 0.06 | boundary | MID|MutS; | 0.625 |
| 45 | fp_beta_D | O15457 | 535 | RGFFIQMTTDCIALPS | EEEEEEEECCCCCCCC | c1a-AT-4 | multi-selected | 0.08 | 0.051 | 0.058 | boundary | MID|MutS; | 0.571 |
| 46 | fp_beta_D | O15457 | 535 | RGFFIQMTTDCIALPS | EEEEEEEECCCCCCCC | c1d-AT-4 | multi | 0.08 | 0.051 | 0.058 | boundary | MID|MutS; | 0.571 |
| 47 | fp_D | O15457 | 537 | FFIQMTTDCIALPS | EEEEEECCCCCCCC | c2-AT-5 | multi | 0.09 | 0.054 | 0.059 | boundary | MID|MutS; | 0.429 |
| 48 | fp_D | O15457 | 565 | NSYSFTSADLIKMNE | CCEEEECHHHHHHHH | c1b-AT-4 | uniq | 0.041 | 0.032 | 0.13 | boundary | MID|MutS; | 0.286 |
| 49 | fp_O | O15457 | 584 | SLREIYHMTYMIVCK | HHHHHHHHHHHHHHH | c1b-5 | multi-selected | 0.041 | 0.012 | 0.042 | ORD | MID|MutS; | 0.0 |
| 50 | fp_O | O15457 | 584 | SLREIYHMTYMIVC | HHHHHHHHHHHHHH | c3-AT | multi-selected | 0.043 | 0.012 | 0.044 | ORD | MID|MutS; | 0.0 |
| 51 | fp_O | O15457 | 592 | TYMIVCKLLSEIYEHI | HHHHHHHHHHHHHHHH | c1aR-4 | uniq | 0.019 | 0.008 | 0.014 | ORD | MID|MutS; | 0.0 |
| 52 | fp_O | O15457 | 599 | LLSEIYEHIHCLYK | HHHHHHHHHHHHHH | c3-4 | uniq | 0.016 | 0.008 | 0.017 | ORD | MID|MutS; | 0.0 |
| 53 | fp_O | O15457 | 606 | HIHCLYKLSDTVSMLD | HHHHHHHHHHHHHHHH | c1d-5 | multi | 0.011 | 0.007 | 0.02 | ORD | MID|MutS; | 0.0 |
| 54 | fp_O | O15457 | 609 | CLYKLSDTVSMLDMLL | HHHHHHHHHHHHHHHH | c1a-5 | multi-selected | 0.01 | 0.007 | 0.022 | ORD | MID|MutS; | 0.0 |
| 55 | fp_O | O15457 | 609 | CLYKLSDTVSMLDMLL | HHHHHHHHHHHHHHHH | c1d-AT-5 | multi | 0.01 | 0.007 | 0.022 | ORD | MID|MutS; | 0.0 |
| 56 | fp_O | O15457 | 612 | KLSDTVSMLDMLLSFAH | HHHHHHHHHHHHHHHHH | c1c-AT-5 | multi | 0.011 | 0.007 | 0.02 | ORD | MID|MutS; | 0.0 |
| 57 | fp_O | O15457 | 612 | KLSDTVSMLDMLLSF | HHHHHHHHHHHHHHH | c1b-AT-5 | multi | 0.011 | 0.006 | 0.02 | ORD | MID|MutS; | 0.0 |
| 58 | fp_O | O15457 | 612 | KLSDTVSMLDMLLS | HHHHHHHHHHHHHH | c3-AT | multi | 0.011 | 0.006 | 0.021 | ORD | MID|MutS; | 0.0 |
| 59 | fp_O | O15457 | 613 | LSDTVSMLDMLLSFAH | HHHHHHHHHHHHHHHH | c1d-4 | multi | 0.011 | 0.007 | 0.02 | ORD | MID|MutS; | 0.0 |
| 60 | fp_O | O15457 | 613 | LSDTVSMLDMLLSF | HHHHHHHHHHHHHH | c2-4 | multi-selected | 0.011 | 0.006 | 0.02 | ORD | MID|MutS; | 0.0 |
| 61 | fp_O | O15457 | 615 | DTVSMLDMLLSFAHAC | HHHHHHHHHHHHHHHH | c1aR-4 | multi-selected | 0.011 | 0.007 | 0.021 | ORD | MID|MutS; | 0.0 |
| 62 | fp_O | O15457 | 632 | LSDYVRPEFTDTLAIKQ | HCCCCCCCCCCEEEECC | c1c-4 | uniq | 0.012 | 0.03 | 0.101 | ORD | MID|MutS; | 0.125 |
| 63 | fp_D | O15457 | 684 | SGKSTYLKQIALCQ | CCHHHHHHHHHHHH | c2-AT-4 | uniq | 0.206 | 0.014 | 0.052 | __ | MID|MutS; | 0.0 |
| 64 | fp_D | O15457 | 691 | KQIALCQIMAQIGSYV | HHHHHHHHHHCCCCCC | c1aR-4 | uniq | 0.066 | 0.008 | 0.03 | boundary | MID|MutS; | 0.0 |
| 65 | fp_D | O15457 | 711 | SSFRIAKQIFTRISTDD | CCCCCCCCCCCCCCCCC | c1cR-4 | multi-selected | 0.064 | 0.03 | 0.141 | boundary | MID|MutS; | 0.0 |
| 66 | fp_D | O15457 | 712 | SFRIAKQIFTRIST | CCCCCCCCCCCCCC | c3-AT | multi-selected | 0.032 | 0.023 | 0.13 | boundary | MID|MutS; | 0.0 |
| 67 | fp_D | O15457 | 736 | FMKEMKEIAYILHN | HHHHHHHHHHHHHC | c3-AT | uniq | 0.279 | 0.016 | 0.145 | boundary | MID|MutS; | 0.0 |
| 68 | fp_O | O15457 | 777 | VCEYLLSLKAFTLFA | HHHHHHCCCCEEEEE | c1b-4 | multi | 0.01 | 0.008 | 0.011 | ORD | MID|MutS; | 0.143 |
| 69 | fp_O | O15457 | 778 | CEYLLSLKAFTLFA | HHHHHCCCCEEEEE | c2-4 | multi-selected | 0.01 | 0.008 | 0.01 | ORD | MID|MutS; | 0.143 |
| 70 | cand_beta_O | O15457 | 783 | SLKAFTLFATHFLELCH * | CCCCEEEEECCCHHHHH | c1c-AT-5 | multi | 0.014 | 0.011 | 0.017 | ORD | MID|MutS; | 0.625 |
| 71 | fp_beta_O | O15457 | 783 | SLKAFTLFATHFLE | CCCCEEEEECCCHH | c3-AT | multi | 0.011 | 0.01 | 0.013 | ORD | MID|MutS; | 0.714 |
| 72 | cand_O | O15457 | 785 | KAFTLFATHFLELCH * | CCEEEEECCCHHHHH | c1b-AT-4 | multi | 0.015 | 0.011 | 0.018 | ORD | MID|MutS; | 0.429 |
| 73 | cand_O | O15457 | 786 | AFTLFATHFLELCH * | CEEEEECCCHHHHH | c2-AT-5 | multi | 0.015 | 0.011 | 0.019 | ORD | MID|MutS; | 0.429 |
| 74 | cand_O | O15457 | 786 | AFTLFATHFLELCH * | CEEEEECCCHHHHH | c3-4 | multi-selected | 0.015 | 0.011 | 0.019 | ORD | MID|MutS; | 0.429 |
| 75 | fp_D | O15457 | 799 | HIDALYPNVENMHFEV * * * | HHCCCCCCEEEEEEEE | c1a-5 | uniq | 0.02 | 0.016 | 0.194 | boundary | MID|MutS; | 0.429 |
| 76 | fp_beta_D | O15457 | 806 | NVENMHFEVQHVKN | CEEEEEEEEEEEEC | c3-4 | uniq | 0.019 | 0.061 | 0.335 | boundary | MID|MutS; | 1.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment