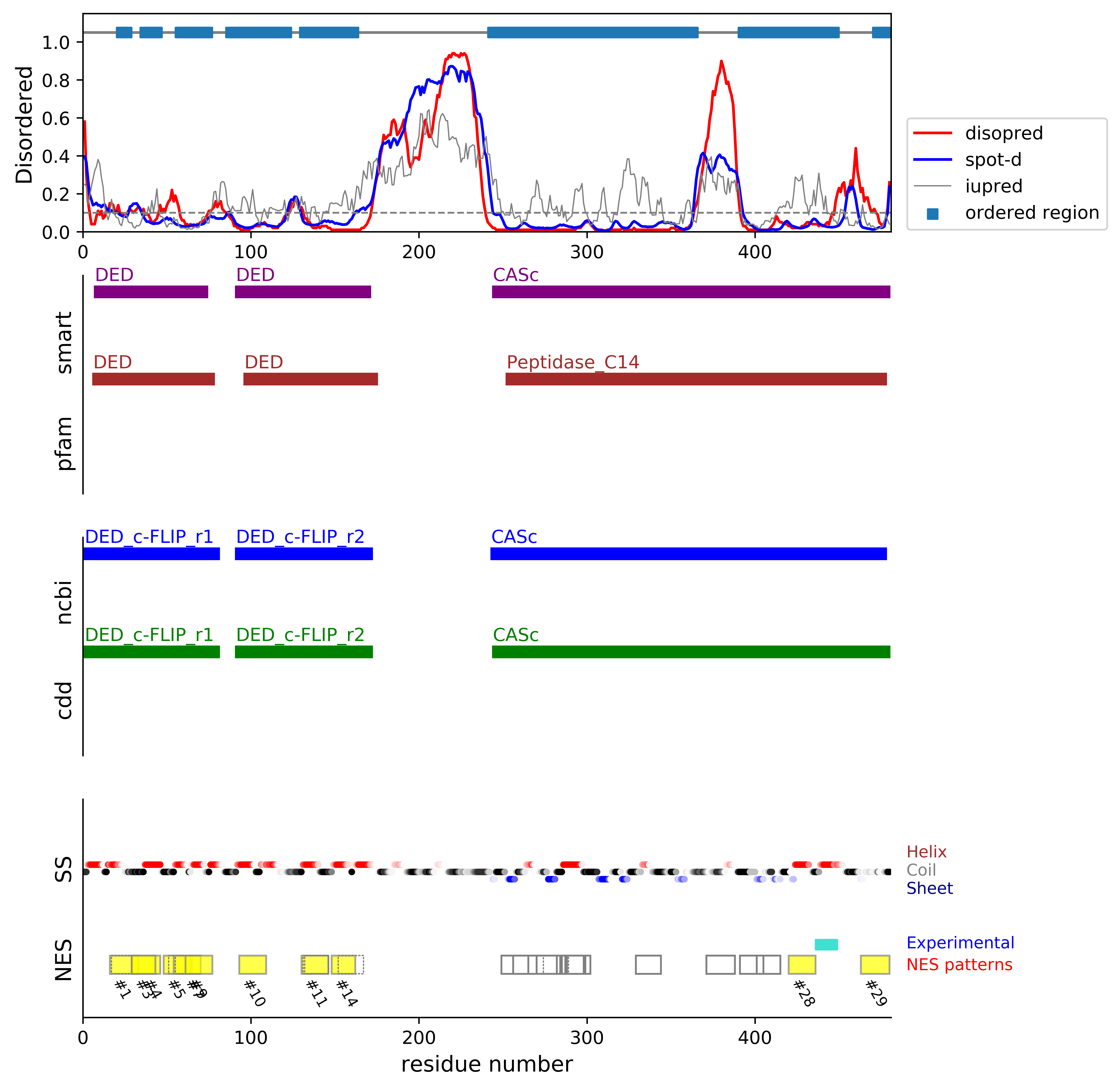

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | O15519 | 16 | DEKEMLLFLCRDVAIDV | HHHHHHHHHCCCCCCCC | c1c-4 | multi-selected | 0.112 | 0.109 | 0.068 | boundary | boundary|DED_c-FLIP_r1; | 0.0 |
| 2 | fp_D | O15519 | 17 | EKEMLLFLCRDVAIDV | HHHHHHHHCCCCCCCC | c1d-4 | multi | 0.11 | 0.109 | 0.067 | boundary | boundary|DED_c-FLIP_r1; | 0.0 |
| 3 | fp_D | O15519 | 29 | AIDVVPPNVRDLLDILR | CCCCCCCCHHHHHHHHH | c1c-5 | multi-selected | 0.093 | 0.086 | 0.09 | boundary | MID|DED_c-FLIP_r1; | 0.0 |
| 4 | fp_D | O15519 | 29 | AIDVVPPNVRDLLD | CCCCCCCCHHHHHH | c3-4 | multi-selected | 0.096 | 0.095 | 0.073 | boundary | MID|DED_c-FLIP_r1; | 0.0 |
| 5 | fp_D | O15519 | 48 | GKLSVGDLAELLYRVR | CCCCCCCHHHHHHHHC | c1aR-5 | multi-selected | 0.117 | 0.046 | 0.038 | boundary | boundary|DED_c-FLIP_r1; | 0.0 |
| 6 | fp_D | O15519 | 51 | SVGDLAELLYRVRR | CCCCHHHHHHHHCC | c3-4 | multi | 0.11 | 0.04 | 0.034 | boundary | boundary|DED_c-FLIP_r1; | 0.0 |
| 7 | fp_D | O15519 | 54 | DLAELLYRVRRFDLLK | CHHHHHHHHCCHHHHH | c1a-5 | multi-selected | 0.073 | 0.034 | 0.029 | boundary | boundary|DED_c-FLIP_r1; | 0.0 |
| 8 | fp_D | O15519 | 55 | LAELLYRVRRFDLLK | HHHHHHHHCCHHHHH | c1b-4 | multi | 0.067 | 0.033 | 0.026 | boundary | boundary|DED_c-FLIP_r1; | 0.0 |
| 9 | fp_D | O15519 | 61 | RVRRFDLLKRILKMDR | HHCCHHHHHHHHCCCH | c1d-5 | uniq | 0.054 | 0.043 | 0.04 | boundary | boundary|DED_c-FLIP_r1; boundary|DED_c-FLIP_r2; | 0.0 |
| 10 | fp_D | O15519 | 93 | YRVLMAEIGEDLDKSD | HHHHHHHHHHCCCHHH | c1aR-4 | uniq | 0.023 | 0.031 | 0.125 | __ | boundary|DED_c-FLIP_r1; boundary|DED_c-FLIP_r2; | 0.0 |
| 11 | fp_D | O15519 | 130 | SFLDLVVELEKLNLVA | CHHHHHHHHHHCCCCC | c1a-5 | multi-selected | 0.046 | 0.047 | 0.106 | boundary | MID|DED_c-FLIP_r2; | 0.0 |
| 12 | fp_D | O15519 | 131 | FLDLVVELEKLNLVA | HHHHHHHHHHCCCCC | c1b-5 | multi | 0.041 | 0.044 | 0.101 | __ | MID|DED_c-FLIP_r2; | 0.0 |
| 13 | fp_D | O15519 | 132 | LDLVVELEKLNLVA | HHHHHHHHHCCCCC | c2-4 | multi | 0.036 | 0.043 | 0.098 | __ | MID|DED_c-FLIP_r2; | 0.0 |
| 14 | fp_D | O15519 | 148 | QLDLLEKCLKNIHR | CHHHHHHHHHHCCH | c3-4 | multi-selected | 0.01 | 0.049 | 0.152 | boundary | boundary|DED_c-FLIP_r2; | 0.0 |
| 15 | fp_D | O15519 | 152 | LEKCLKNIHRIDLKT | HHHHHHHCCHHHHHH | c1b-4 | multi | 0.011 | 0.07 | 0.172 | boundary | boundary|DED_c-FLIP_r2; | 0.0 |
| 16 | fp_beta_D | O15519 | 249 | KPLGICLIIDCIGNET | CCCEEEEEECCCCCCH | c1aR-4 | uniq | 0.01 | 0.032 | 0.084 | boundary | boundary|CASc; | 0.714 |
| 17 | fp_D | O15519 | 256 | IIDCIGNETELLRD | EECCCCCCHHHHHH | c3-AT | uniq | 0.01 | 0.029 | 0.099 | boundary | boundary|CASc; | 0.0 |
| 18 | fp_beta_O | O15519 | 270 | TFTSLGYEVQKFLHLSM | HCCCCCCEEEEECCCCH | c1c-5 | multi-selected | 0.01 | 0.024 | 0.138 | ORD | MID|CASc; | 0.625 |
| 19 | fp_O | O15519 | 270 | TFTSLGYEVQKFLH | HCCCCCCEEEEECC | c3-4 | multi-selected | 0.01 | 0.021 | 0.146 | ORD | MID|CASc; | 0.429 |
| 20 | fp_beta_O | O15519 | 274 | LGYEVQKFLHLSMHG | CCCEEEEECCCCHHH | c1b-4 | multi | 0.01 | 0.026 | 0.109 | ORD | MID|CASc; | 0.571 |
| 21 | fp_O | O15519 | 282 | LHLSMHGISQILGQFA | CCCCHHHHHHHHHHHH | c1aR-5 | multi-selected | 0.013 | 0.029 | 0.137 | ORD | MID|CASc; | 0.0 |
| 22 | fp_O | O15519 | 285 | SMHGISQILGQFAC | CHHHHHHHHHHHHC | c3-4 | multi-selected | 0.016 | 0.029 | 0.146 | ORD | MID|CASc; | 0.0 |
| 23 | fp_O | O15519 | 288 | GISQILGQFACMPE | HHHHHHHHHHCCCC | c3-4 | multi-selected | 0.025 | 0.034 | 0.142 | ORD | MID|CASc; | 0.0 |
| 24 | fp_O | O15519 | 329 | SGLPLHHIRRMFMGD | CCCCHHHHHCCCCCC | c1b-4 | uniq | 0.01 | 0.033 | 0.178 | ORD | MID|CASc; | 0.0 |
| 25 | fp_D | O15519 | 371 | SLLEVDGPAMKNVEFKA | CCCCCCCCCCCCHHHHH | c1c-AT-5 | uniq | 0.729 | 0.357 | 0.283 | boundary | MID|CASc; | 0.0 |
| 26 | fp_D | O15519 | 391 | GLCTVHREADFFWS | CCCCCCCCCCEEEE | c3-AT | uniq | 0.024 | 0.056 | 0.051 | boundary | MID|CASc; | 0.0 |
| 27 | fp_D | O15519 | 401 | FFWSLCTADMSLLE | EEEEECCCCCEEEE | c2-AT-5 | uniq | 0.026 | 0.011 | 0.058 | boundary | MID|CASc; | 0.286 |
| 28 | fp_D | O15519 | 420 | PSLYLQCLSQKLRQER | CCEEHHHHHHHHHCCC | c1aR-4 | uniq | 0.032 | 0.033 | 0.194 | boundary | MID|CASc; | 0.0 |
| 29 | fp_D | O15519 | 463 | YYVWLQHTLRKKLILSY | EECCCCCCCCCCCCCCC | c1c-4 | uniq | 0.118 | 0.028 | 0.048 | boundary | boundary|CASc; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment