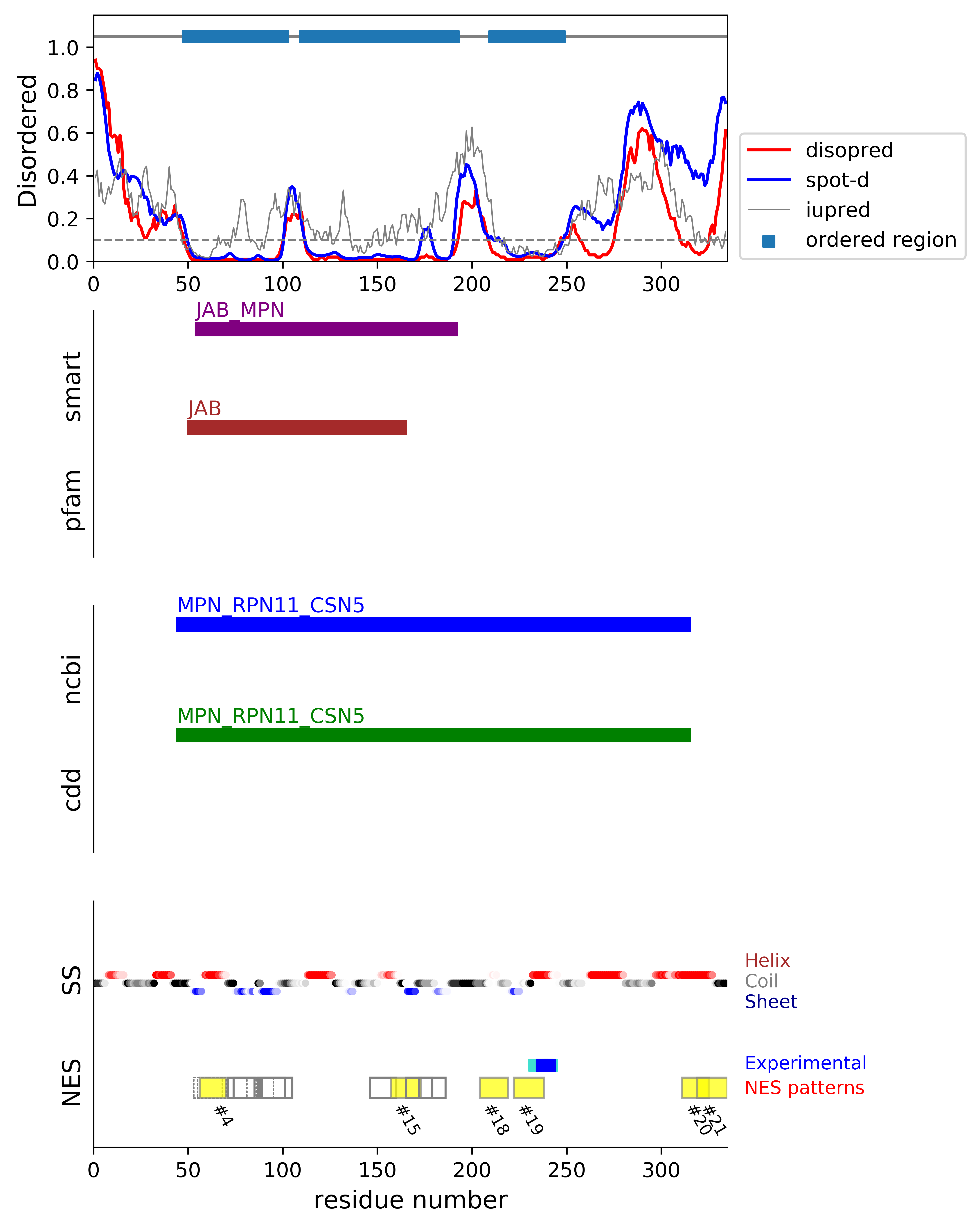

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | O35864 | 53 | KYCKISALALLKMVMHA | EEEEEHHHHHHHHHHHH | c1c-AT-4 | multi | 0.009 | 0.016 | 0.049 | boundary | boundary|MPN_RPN11_CSN5; | 0.125 |
| 2 | fp_D | O35864 | 53 | KYCKISALALLKMVM | EEEEEHHHHHHHHHH | c1b-4 | multi | 0.009 | 0.015 | 0.042 | boundary | boundary|MPN_RPN11_CSN5; | 0.143 |
| 3 | fp_D | O35864 | 55 | CKISALALLKMVMHA | EEEHHHHHHHHHHHH | c1b-AT-4 | multi | 0.009 | 0.015 | 0.05 | boundary | boundary|MPN_RPN11_CSN5; | 0.0 |
| 4 | fp_D | O35864 | 56 | KISALALLKMVMHA | EEHHHHHHHHHHHH | c2-5 | multi-selected | 0.009 | 0.015 | 0.052 | boundary | boundary|MPN_RPN11_CSN5; | 0.0 |
| 5 | fp_beta_O | O35864 | 71 | SGGNLEVMGLMLGK | CCCCEEEEEEEEEE | c2-4 | multi-selected | 0.01 | 0.016 | 0.161 | ORD | MID|MPN_RPN11_CSN5; | 0.857 |
| 6 | fp_beta_D | O35864 | 74 | NLEVMGLMLGKVDG | CEEEEEEEEEEEEC | c3-4 | multi-selected | 0.01 | 0.013 | 0.159 | boundary | MID|MPN_RPN11_CSN5; | 1.0 |
| 7 | fp_beta_D | O35864 | 81 | MLGKVDGETMIIMD | EEEEEECCEEEEEE | c3-AT | multi | 0.01 | 0.013 | 0.118 | boundary | MID|MPN_RPN11_CSN5; | 0.714 |
| 8 | fp_beta_D | O35864 | 85 | VDGETMIIMDSFALPV | EECCEEEEEEEEECCC | c1a-AT-4 | multi | 0.02 | 0.017 | 0.165 | boundary | MID|MPN_RPN11_CSN5; | 1.0 |
| 9 | fp_beta_D | O35864 | 85 | VDGETMIIMDSFALPV | EECCEEEEEEEEECCC | c1d-AT-4 | multi | 0.02 | 0.017 | 0.165 | boundary | MID|MPN_RPN11_CSN5; | 1.0 |
| 10 | fp_beta_D | O35864 | 86 | DGETMIIMDSFALPV | ECCEEEEEEEEECCC | c1b-4 | multi | 0.021 | 0.017 | 0.169 | boundary | MID|MPN_RPN11_CSN5; | 1.0 |
| 11 | fp_beta_D | O35864 | 87 | GETMIIMDSFALPV | CCEEEEEEEEECCC | c2-4 | multi-selected | 0.021 | 0.016 | 0.176 | boundary | MID|MPN_RPN11_CSN5; | 1.0 |
| 12 | fp_beta_D | O35864 | 88 | ETMIIMDSFALPVEGTE | CEEEEEEEEECCCCCCC | c1cR-4 | multi | 0.059 | 0.074 | 0.21 | boundary | MID|MPN_RPN11_CSN5; | 0.75 |
| 13 | fp_beta_D | O35864 | 89 | TMIIMDSFALPVEGTE | EEEEEEEEECCCCCCC | c1aR-4 | multi-selected | 0.062 | 0.077 | 0.22 | boundary | MID|MPN_RPN11_CSN5; | 0.714 |
| 14 | fp_O | O35864 | 146 | WLSGIDVSTQMLNQ | CCCCCHHHHHHHHH | c3-AT | uniq | 0.01 | 0.024 | 0.115 | ORD | MID|MPN_RPN11_CSN5; | 0.0 |
| 15 | fp_D | O35864 | 157 | LNQQFQEPFVAVVIDP | HHHHHCCCCEEEEECC | c1a-4 | uniq | 0.01 | 0.018 | 0.175 | boundary | MID|MPN_RPN11_CSN5; | 0.286 |
| 16 | fp_D | O35864 | 165 | FVAVVIDPTRTISA | CEEEEECCCCCCCC | c3-AT | uniq | 0.015 | 0.066 | 0.171 | boundary | MID|MPN_RPN11_CSN5; | 0.429 |
| 17 | fp_D | O35864 | 172 | PTRTISAGKVNLGA | CCCCCCCCCEEEEE | c2-AT-4 | uniq | 0.016 | 0.077 | 0.195 | boundary | MID|MPN_RPN11_CSN5; | 0.143 |
| 18 | fp_D | O35864 | 204 | QTIPLNKIEDFGVHC | CCCCCCHHHHCCCCC | c1b-4 | uniq | 0.085 | 0.122 | 0.236 | boundary | MID|MPN_RPN11_CSN5; | 0.0 |
| 19 | fp_D | O35864 | 222 | YALEVSYFKSSLDRKL ++++* | EEEECCCCCCHHHHHH | c1aR-4 | uniq | 0.016 | 0.032 | 0.046 | boundary | MID|MPN_RPN11_CSN5; | 0.0 |
| 20 | fp_D | O35864 | 311 | TIEAIHGLMSQVIK | HHHHHHHHHHHHHH | c3-4 | uniq | 0.058 | 0.425 | 0.159 | DISO | boundary|MPN_RPN11_CSN5; | 0.0 |
| 21 | fp_D | O35864 | 319 | MSQVIKDKLFNQINVAX | HHHHHHHHHCCCCCCC | c1c-4 | uniq | 0.196 | 0.529 | 0.101 | DISO | boundary|MPN_RPN11_CSN5; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment