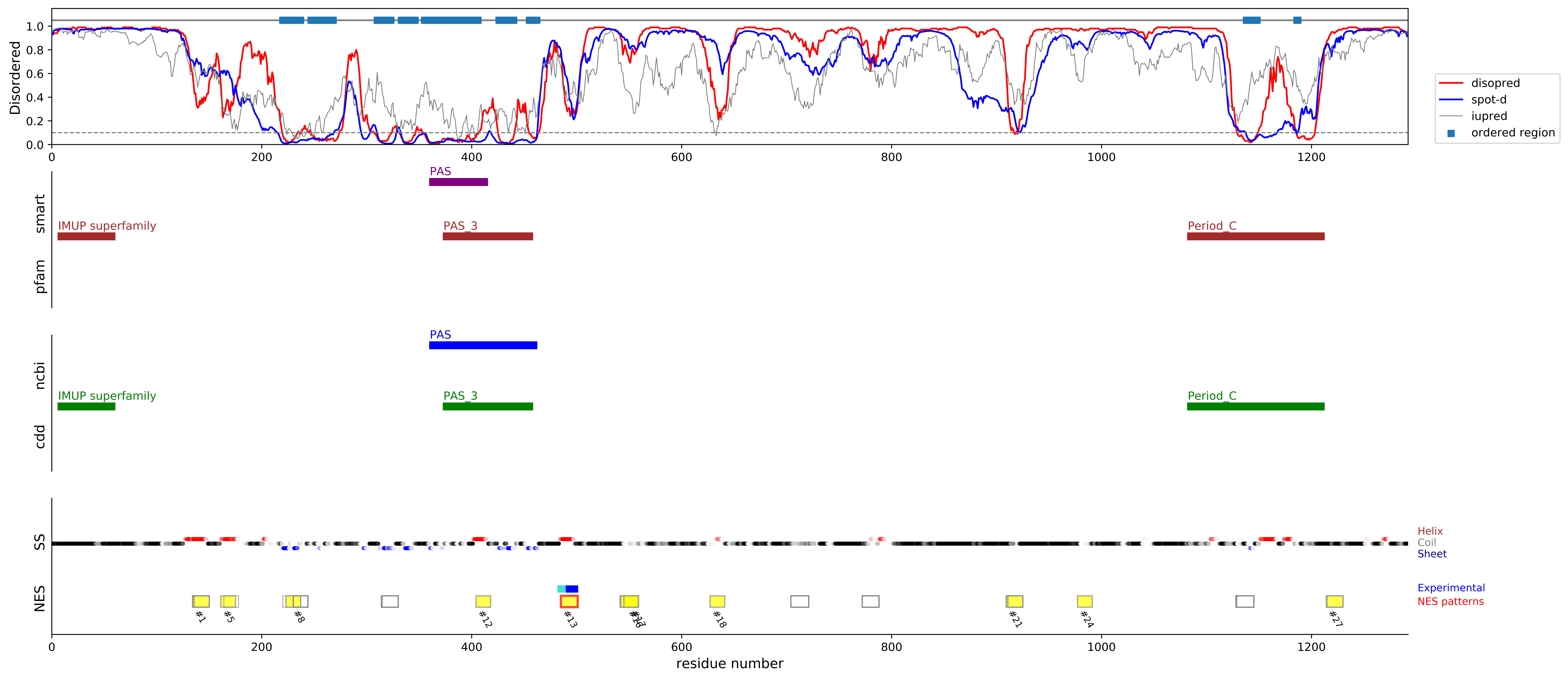

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | O35973 | 134 | TQKELMTALRELKLRL | HHHHHHHHHHHHHHCC | c1a-4 | multi-selected | 0.402 | 0.65 | 0.6 | DISO | 0.0 | |
| 2 | fp_D | O35973 | 134 | TQKELMTALRELKLRL | HHHHHHHHHHHHHHCC | c1d-AT-4 | multi | 0.402 | 0.65 | 0.6 | DISO | 0.0 | |
| 3 | fp_D | O35973 | 135 | QKELMTALRELKLRL | HHHHHHHHHHHHHCC | c1b-4 | multi | 0.388 | 0.646 | 0.599 | DISO | 0.0 | |
| 4 | fp_D | O35973 | 136 | KELMTALRELKLRL | HHHHHHHHHHHHCC | c2-AT-4 | multi | 0.379 | 0.643 | 0.594 | DISO | 0.0 | |
| 5 | fp_D | O35973 | 161 | TLATLQYALACVKQ | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.341 | 0.572 | 0.26 | DISO | 0.0 | |
| 6 | fp_D | O35973 | 164 | TLQYALACVKQVQA | HHHHHHHHHHHHHH | c3-AT | multi | 0.368 | 0.536 | 0.215 | DISO | 0.0 | |
| 7 | fp_beta_D | O35973 | 220 | FSVAVSFLTGRIVYISE | EEEEEEECCCCEEEECC | c1c-AT-4 | multi | 0.052 | 0.017 | 0.109 | boundary | 0.5 | |
| 8 | fp_D | O35973 | 223 | AVSFLTGRIVYISE | EEEECCCCEEEECC | c3-4 | multi-selected | 0.051 | 0.018 | 0.09 | boundary | 0.429 | |
| 9 | fp_D | O35973 | 230 | RIVYISEQAGVLLR | CEEEECCCHHHCCC | c3-AT | uniq | 0.1 | 0.029 | 0.08 | boundary | 0.286 | |
| 10 | fp_beta_D | O35973 | 314 | QPFRLTPYVTKIRVSD | EEEEEEEEEEEEECCC | c1a-4 | multi-selected | 0.079 | 0.026 | 0.306 | boundary | 1.0 | |
| 11 | fp_beta_D | O35973 | 315 | PFRLTPYVTKIRVSD | EEEEEEEEEEEECCC | c1b-AT-5 | multi | 0.082 | 0.027 | 0.291 | boundary | 1.0 | |
| 12 | fp_D | O35973 | 404 | LMLAIHKKILQLAG | HHHHHHHHHHHCCC | c3-4 | uniq | 0.198 | 0.047 | 0.198 | boundary | MID|PAS_3; | 0.0 |
| 13 | cand_D | O35973 | 485 | DIQELSEQIHRLLLQP ++++++++*++*+* | HHHHHHHHHHHHHCCC | c1a-5 | uniq | 0.369 | 0.465 | 0.461 | DISO | boundary|PAS_3; | 0.0 |
| 14 | fp_D | O35973 | 541 | PAPVTFQQICKDVHLVK | CCCCCCCCCCCCCCCCC | c1c-AT-4 | multi | 0.795 | 0.864 | 0.458 | DISO | 0.0 | |
| 15 | fp_D | O35973 | 542 | APVTFQQICKDVHLVK | CCCCCCCCCCCCCCCC | c1d-4 | multi | 0.787 | 0.859 | 0.444 | DISO | 0.0 | |
| 16 | fp_D | O35973 | 542 | APVTFQQICKDVHLVK | CCCCCCCCCCCCCCCC | c1aR-5 | multi-selected | 0.787 | 0.859 | 0.444 | DISO | 0.0 | |
| 17 | fp_D | O35973 | 545 | TFQQICKDVHLVKH | CCCCCCCCCCCCCC | c3-4 | multi-selected | 0.776 | 0.843 | 0.395 | DISO | 0.0 | |
| 18 | fp_D | O35973 | 627 | QINCLDSILRYLES | CCCCHHHHHHHHHC | c3-4 | uniq | 0.371 | 0.785 | 0.186 | DISO | 0.0 | |
| 19 | fp_D | O35973 | 704 | SPLALANKAESVVSVTS | CCCCCCCCCCCCCCCCC | c1c-AT-4 | uniq | 0.881 | 0.73 | 0.401 | DISO | 0.0 | |
| 20 | fp_D | O35973 | 772 | AYRPVGLTKAVLSLHT | CCCCCCCHHHHHHCCC | c1d-AT-4 | uniq | 0.774 | 0.713 | 0.623 | DISO | 0.0 | |
| 21 | fp_D | O35973 | 909 | FPSPLVTPMVALVLPN | CCCCCCCCEEECCCCC | c1a-4 | multi-selected | 0.212 | 0.218 | 0.472 | DISO | 0.429 | |
| 22 | fp_D | O35973 | 910 | PSPLVTPMVALVLPN | CCCCCCCEEECCCCC | c1b-4 | multi | 0.184 | 0.212 | 0.459 | DISO | 0.429 | |
| 23 | fp_D | O35973 | 911 | SPLVTPMVALVLPN | CCCCCCEEECCCCC | c2-AT-4 | multi | 0.161 | 0.207 | 0.448 | DISO | 0.429 | |
| 24 | fp_D | O35973 | 977 | CSSPLQLNLLQLEE | CCCCCCCCCCCCCC | c2-4 | uniq | 0.971 | 0.842 | 0.667 | DISO | 0.0 | |
| 25 | fp_D | O35973 | 1128 | PIWLLMANADQRVMMTY | CCCHHCCCCCCCCEEEE | c1c-AT-5 | multi-selected | 0.054 | 0.09 | 0.362 | boundary | MID|Period_C; | 0.0 |
| 26 | fp_D | O35973 | 1129 | IWLLMANADQRVMMTY | CCHHCCCCCCCCEEEE | c1d-AT-4 | multi | 0.048 | 0.086 | 0.368 | boundary | MID|Period_C; | 0.0 |
| 27 | fp_D | O35973 | 1214 | SDDPLFSELDGLGLEP | CCCCCCCCCCCCCCCC | c1a-4 | multi-selected | 0.938 | 0.848 | 0.771 | DISO | boundary|Period_C; | 0.0 |
| 28 | fp_D | O35973 | 1215 | DDPLFSELDGLGLEP | CCCCCCCCCCCCCCC | c1b-4 | multi | 0.944 | 0.849 | 0.773 | DISO | boundary|Period_C; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment