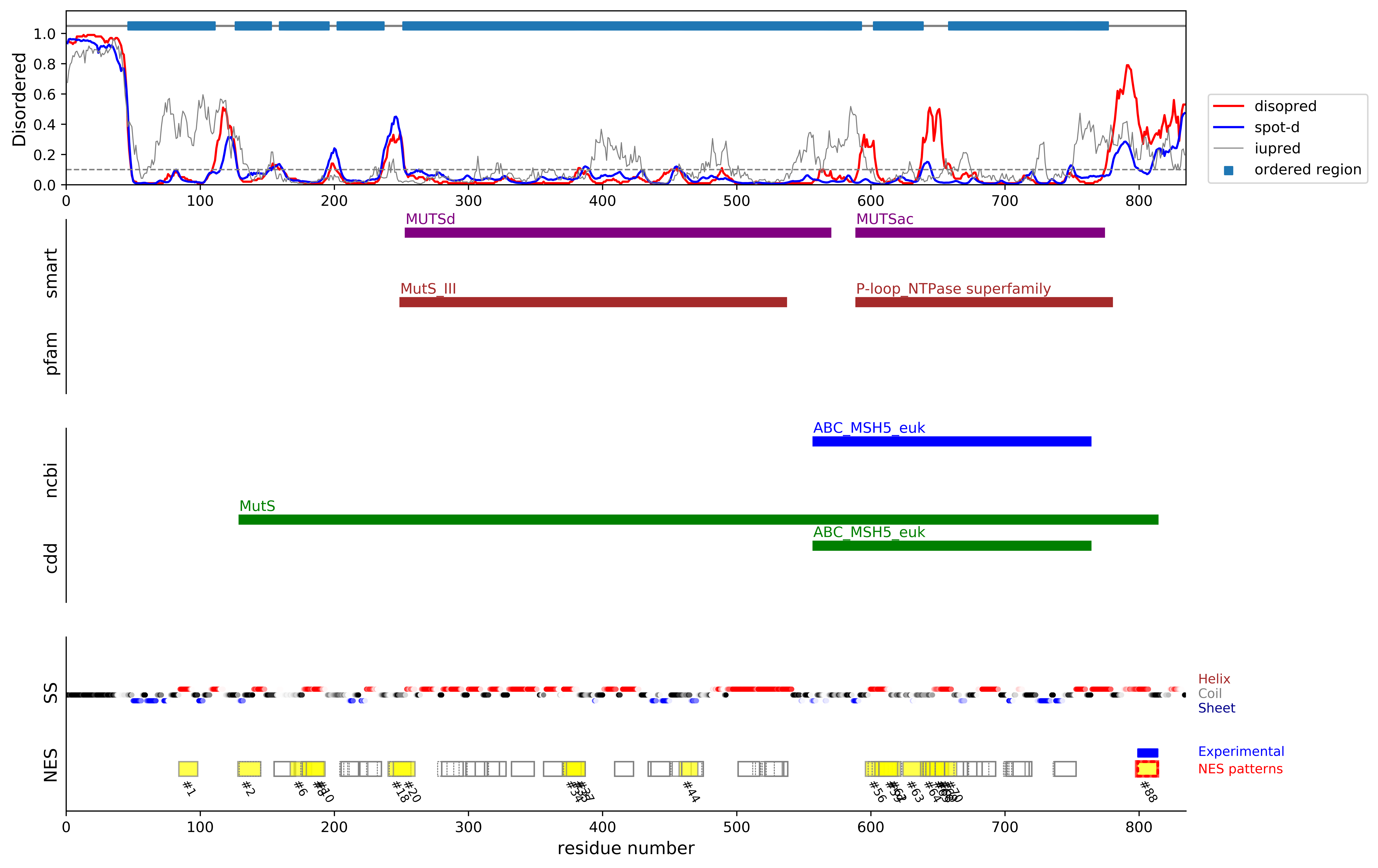

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | O43196 | 84 | SLKLLQRVLDEINP | CHHHHHHHHHHCCC | c3-4 | uniq | 0.036 | 0.029 | 0.367 | boundary | 0.0 | |
| 2 | fp_D | O43196 | 128 | PEIIFLPSVDFGLEISK | CEEEECCCCCCCHHHHH | c1c-4 | multi-selected | 0.034 | 0.063 | 0.129 | boundary | boundary|MutS; | 0.125 |
| 3 | fp_D | O43196 | 129 | EIIFLPSVDFGLEISK | EEEECCCCCCCHHHHH | c1d-5 | multi | 0.029 | 0.061 | 0.119 | boundary | boundary|MutS; | 0.0 |
| 4 | fp_D | O43196 | 155 | IPDAMTATEKILFLS | CCCCHHCCCCCCCCC | c1b-AT-4 | multi-selected | 0.083 | 0.084 | 0.048 | boundary | MID|MutS; | 0.0 |
| 5 | fp_D | O43196 | 155 | IPDAMTATEKILFLSS | CCCCHHCCCCCCCCCC | c1d-AT-4 | multi-selected | 0.081 | 0.081 | 0.046 | boundary | MID|MutS; | 0.0 |
| 6 | fp_D | O43196 | 167 | FLSSIIPFDCLLTVRA | CCCCCCCCCCHHHHHH | c1d-5 | uniq | 0.024 | 0.025 | 0.013 | boundary | MID|MutS; | 0.0 |
| 7 | fp_D | O43196 | 175 | DCLLTVRALGGLLKFLG | CCHHHHHHHHHHHHHHH | c1c-AT-4 | multi | 0.011 | 0.021 | 0.021 | boundary | MID|MutS; | 0.0 |
| 8 | fp_D | O43196 | 176 | CLLTVRALGGLLKFLG | CHHHHHHHHHHHHHHH | c1aR-5 | multi-selected | 0.011 | 0.021 | 0.021 | boundary | MID|MutS; | 0.0 |
| 9 | fp_D | O43196 | 176 | CLLTVRALGGLLKFLG | CHHHHHHHHHHHHHHH | c1d-5 | multi | 0.011 | 0.021 | 0.021 | boundary | MID|MutS; | 0.0 |
| 10 | fp_D | O43196 | 179 | TVRALGGLLKFLGR | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.011 | 0.023 | 0.024 | boundary | MID|MutS; | 0.0 |
| 11 | fp_beta_D | O43196 | 204 | VSVPILGFKKFMLTH | CCCCCCCEEEECCCC | c1b-4 | multi | 0.02 | 0.042 | 0.02 | boundary | MID|MutS; | 0.571 |
| 12 | fp_beta_D | O43196 | 205 | SVPILGFKKFMLTH | CCCCCCEEEECCCC | c2-5 | multi-selected | 0.016 | 0.037 | 0.02 | boundary | MID|MutS; | 0.571 |
| 13 | fp_beta_D | O43196 | 207 | PILGFKKFMLTHLVNID | CCCCEEEECCCCEEEEE | c1cR-5 | multi | 0.011 | 0.027 | 0.02 | boundary | MID|MutS; | 0.5 |
| 14 | fp_D | O43196 | 210 | GFKKFMLTHLVNIDQ | CEEEECCCCEEEEEH | c1b-AT-5 | multi | 0.01 | 0.025 | 0.019 | boundary | MID|MutS; | 0.429 |
| 15 | fp_D | O43196 | 211 | FKKFMLTHLVNIDQ | EEEECCCCEEEEEH | c2-AT-4 | multi | 0.01 | 0.024 | 0.019 | boundary | MID|MutS; | 0.429 |
| 16 | fp_D | O43196 | 218 | HLVNIDQDTYSVLQIFK | CEEEEEHHHHHHHHHCC | c1c-AT-5 | multi-selected | 0.015 | 0.032 | 0.049 | boundary | MID|MutS; | 0.25 |
| 17 | fp_D | O43196 | 218 | HLVNIDQDTYSVLQ | CEEEEEHHHHHHHH | c3-AT | multi | 0.011 | 0.028 | 0.043 | boundary | MID|MutS; | 0.429 |
| 18 | fp_D | O43196 | 240 | SVYKVASGLKEGLSLFG | CCCCCCCCCCCCCCHHH | c1c-5 | multi-selected | 0.229 | 0.278 | 0.06 | __ | MID|MutS; | 0.0 |
| 19 | fp_D | O43196 | 241 | VYKVASGLKEGLSLFG | CCCCCCCCCCCCCHHH | c1d-AT-4 | multi | 0.228 | 0.276 | 0.059 | boundary | MID|MutS; | 0.0 |
| 20 | fp_D | O43196 | 244 | VASGLKEGLSLFGILN | CCCCCCCCCCHHHHHH | c1a-4 | multi-selected | 0.186 | 0.223 | 0.04 | boundary | MID|MutS; | 0.0 |
| 21 | fp_O | O43196 | 277 | PTHDLGELSSRLDVIQ | HCCCHHHHHHHHHHHH | c1d-4 | multi | 0.013 | 0.046 | 0.042 | ORD | MID|MutS; | 0.0 |
| 22 | fp_O | O43196 | 280 | DLGELSSRLDVIQFFL | CHHHHHHHHHHHHHHH | c1a-5 | multi-selected | 0.013 | 0.038 | 0.036 | ORD | MID|MutS; | 0.0 |
| 23 | fp_O | O43196 | 284 | LSSRLDVIQFFLLP | HHHHHHHHHHHHCC | c2-4 | multi | 0.011 | 0.033 | 0.029 | ORD | MID|MutS; | 0.0 |
| 24 | fp_O | O43196 | 284 | LSSRLDVIQFFLLPQ | HHHHHHHHHHHHCCC | c1b-4 | multi | 0.011 | 0.034 | 0.028 | ORD | MID|MutS; | 0.0 |
| 25 | fp_O | O43196 | 289 | DVIQFFLLPQNLDMAQ | HHHHHHHCCCCHHHHH | c1aR-4 | multi | 0.01 | 0.039 | 0.022 | ORD | MID|MutS; | 0.0 |
| 26 | fp_O | O43196 | 298 | QNLDMAQMLHRLLGHIK | CCHHHHHHHHHHCCCCC | c1cR-5 | multi | 0.011 | 0.04 | 0.052 | ORD | MID|MutS; | 0.0 |
| 27 | fp_O | O43196 | 298 | QNLDMAQMLHRLLGHI | CCHHHHHHHHHHCCCC | c1aR-4 | multi-selected | 0.011 | 0.04 | 0.051 | ORD | MID|MutS; | 0.0 |
| 28 | fp_O | O43196 | 305 | MLHRLLGHIKNVPLILKR | HHHHHCCCCCCHHHHHHH | c4-5 | uniq | 0.013 | 0.036 | 0.078 | ORD | MID|MutS; | 0.0 |
| 29 | fp_O | O43196 | 312 | HIKNVPLILKRMKLSH | CCCCHHHHHHHHHCCC | c1a-5 | multi-selected | 0.021 | 0.041 | 0.076 | ORD | MID|MutS; | 0.0 |
| 30 | fp_O | O43196 | 312 | HIKNVPLILKRMKLSH | CCCCHHHHHHHHHCCC | c1d-5 | multi | 0.021 | 0.041 | 0.076 | ORD | MID|MutS; | 0.0 |
| 31 | fp_O | O43196 | 314 | KNVPLILKRMKLSH | CCHHHHHHHHHCCC | c2-4 | multi | 0.023 | 0.041 | 0.08 | ORD | MID|MutS; | 0.0 |
| 32 | fp_O | O43196 | 332 | DWQVLYKTVYSALGLRD | HHHHHHHHHHHHHHHHH | c1c-4 | uniq | 0.017 | 0.014 | 0.047 | ORD | MID|MutS; | 0.0 |
| 33 | fp_D | O43196 | 356 | SIQLFRDIAQEFSD | CHHHHHHHHHHHCC | c3-AT | uniq | 0.011 | 0.039 | 0.075 | boundary | MID|MutS; | 0.0 |
| 34 | fp_D | O43196 | 370 | DLHHIASLIGKVVDFEG | CHHHHHHHHHHHCCCCC | c1c-5 | multi-selected | 0.048 | 0.051 | 0.052 | boundary | MID|MutS; | 0.0 |
| 35 | fp_D | O43196 | 370 | DLHHIASLIGKVVD | CHHHHHHHHHHHCC | c3-4 | multi-selected | 0.04 | 0.041 | 0.054 | boundary | MID|MutS; | 0.0 |
| 36 | fp_D | O43196 | 371 | LHHIASLIGKVVDFEG | HHHHHHHHHHHCCCCC | c1d-AT-4 | multi | 0.05 | 0.051 | 0.052 | boundary | MID|MutS; | 0.0 |
| 37 | fp_D | O43196 | 373 | HIASLIGKVVDFEG | HHHHHHHHHCCCCC | c3-4 | multi-selected | 0.056 | 0.052 | 0.05 | boundary | MID|MutS; | 0.0 |
| 38 | fp_O | O43196 | 409 | RLMGLPSFLTEVAR | HHCCCHHHHHHHHH | c3-4 | uniq | 0.024 | 0.058 | 0.187 | ORD | MID|MutS; | 0.0 |
| 39 | fp_beta_D | O43196 | 434 | SCSVIYIPLIGFLLSI | CCEEEEECCCEEEEEE | c1a-4 | multi-selected | 0.04 | 0.012 | 0.014 | boundary | MID|MutS; | 0.571 |
| 40 | fp_beta_D | O43196 | 436 | SVIYIPLIGFLLSI | EEEEECCCEEEEEE | c2-5 | multi-selected | 0.043 | 0.01 | 0.012 | boundary | MID|MutS; | 0.571 |
| 41 | fp_beta_D | O43196 | 436 | SVIYIPLIGFLLSIPR | EEEEECCCEEEEEEEC | c1d-5 | multi | 0.049 | 0.013 | 0.015 | boundary | MID|MutS; | 0.571 |
| 42 | fp_D | O43196 | 451 | RLPSMVEASDFEING | CCCCCCCCCCCCCCC | c1b-AT-5 | multi | 0.068 | 0.081 | 0.069 | boundary | MID|MutS; | 0.0 |
| 43 | fp_D | O43196 | 452 | LPSMVEASDFEING | CCCCCCCCCCCCCC | c2-AT-4 | multi | 0.066 | 0.084 | 0.071 | boundary | MID|MutS; | 0.0 |
| 44 | fp_D | O43196 | 457 | EASDFEINGLDFMF | CCCCCCCCCEEEEE | c2-4 | multi-selected | 0.034 | 0.054 | 0.073 | boundary | MID|MutS; | 0.143 |
| 45 | fp_beta_D | O43196 | 459 | SDFEINGLDFMFLSEE | CCCCCCCEEEEECCCC | c1aR-4 | multi-selected | 0.02 | 0.042 | 0.069 | __ | MID|MutS; | 0.571 |
| 46 | fp_beta_D | O43196 | 459 | SDFEINGLDFMFLSE | CCCCCCCEEEEECCC | c1b-4 | multi | 0.021 | 0.042 | 0.064 | __ | MID|MutS; | 0.571 |
| 47 | fp_O | O43196 | 501 | ETLLMYQLQCQVLARA | HHHHHHHHHHHHHHHH | c1aR-4 | uniq | 0.011 | 0.012 | 0.05 | ORD | MID|MutS; | 0.0 |
| 48 | fp_O | O43196 | 512 | VLARAAVLTRVLDLAS | HHHHHHHHHHHHHHHH | c1d-AT-5 | multi | 0.011 | 0.011 | 0.023 | ORD | MID|MutS; | 0.0 |
| 49 | fp_O | O43196 | 514 | ARAAVLTRVLDLAS | HHHHHHHHHHHHHH | c2-AT-4 | multi | 0.011 | 0.011 | 0.024 | ORD | MID|MutS; | 0.0 |
| 50 | fp_O | O43196 | 518 | VLTRVLDLASRLDVLL | HHHHHHHHHHHHHHHH | c1a-AT-5 | multi | 0.01 | 0.008 | 0.025 | ORD | MID|MutS; | 0.0 |
| 51 | fp_O | O43196 | 518 | VLTRVLDLASRLDVLL | HHHHHHHHHHHHHHHH | c1d-5 | multi | 0.01 | 0.008 | 0.025 | ORD | MID|MutS; | 0.0 |
| 52 | fp_O | O43196 | 519 | LTRVLDLASRLDVLL | HHHHHHHHHHHHHHH | c1b-AT-4 | multi | 0.01 | 0.008 | 0.024 | ORD | MID|MutS; | 0.0 |
| 53 | fp_O | O43196 | 521 | RVLDLASRLDVLLALAS | HHHHHHHHHHHHHHHHH | c1c-5 | multi-selected | 0.01 | 0.008 | 0.028 | ORD | MID|MutS; boundary|ABC_MSH5_euk; | 0.0 |
| 54 | fp_O | O43196 | 521 | RVLDLASRLDVLLA | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.01 | 0.007 | 0.024 | ORD | MID|MutS; | 0.0 |
| 55 | fp_O | O43196 | 522 | VLDLASRLDVLLALAS | HHHHHHHHHHHHHHHH | c1d-AT-5 | multi | 0.01 | 0.008 | 0.028 | ORD | MID|MutS; boundary|ABC_MSH5_euk; | 0.0 |
| 56 | fp_D | O43196 | 596 | SGKSIYLKQVGLIT | CCCHHHHHHHHHHH | c2-4 | multi-selected | 0.17 | 0.015 | 0.066 | __ | MID|MutS; MID|ABC_MSH5_euk; | 0.0 |
| 57 | fp_D | O43196 | 598 | KSIYLKQVGLITFMA | CHHHHHHHHHHHHHH | c1b-4 | multi | 0.125 | 0.011 | 0.046 | boundary | MID|MutS; MID|ABC_MSH5_euk; | 0.0 |
| 58 | fp_D | O43196 | 601 | YLKQVGLITFMALVG | HHHHHHHHHHHHHHC | c1b-5 | multi | 0.079 | 0.009 | 0.035 | boundary | MID|MutS; MID|ABC_MSH5_euk; | 0.0 |
| 59 | fp_D | O43196 | 603 | KQVGLITFMALVGSFV | HHHHHHHHHHHHCCCC | c1aR-5 | multi-selected | 0.046 | 0.011 | 0.026 | boundary | MID|MutS; MID|ABC_MSH5_euk; | 0.0 |
| 60 | fp_D | O43196 | 606 | GLITFMALVGSFVPAEE | HHHHHHHHHCCCCCCCC | c1cR-4 | multi | 0.029 | 0.019 | 0.031 | boundary | MID|MutS; MID|ABC_MSH5_euk; | 0.0 |
| 61 | fp_D | O43196 | 606 | GLITFMALVGSFVPAE | HHHHHHHHHCCCCCCC | c1aR-4 | multi-selected | 0.029 | 0.017 | 0.031 | boundary | MID|MutS; MID|ABC_MSH5_euk; | 0.0 |
| 62 | fp_D | O43196 | 606 | GLITFMALVGSFVP | HHHHHHHHHCCCCC | c3-4 | multi-selected | 0.029 | 0.013 | 0.028 | boundary | MID|MutS; MID|ABC_MSH5_euk; | 0.0 |
| 63 | fp_D | O43196 | 624 | EIGAVDAIFTRIHS | CCCCCCCEECCCCC | c3-4 | uniq | 0.03 | 0.032 | 0.076 | boundary | MID|MutS; MID|ABC_MSH5_euk; | 0.286 |
| 64 | fp_D | O43196 | 637 | SCESISLGLSTFMIDL | CCCCCCCCCCHHHHHH | c1a-4 | multi-selected | 0.373 | 0.082 | 0.044 | boundary | MID|MutS; MID|ABC_MSH5_euk; | 0.0 |
| 65 | fp_D | O43196 | 639 | ESISLGLSTFMIDL | CCCCCCCCHHHHHH | c2-4 | multi-selected | 0.405 | 0.082 | 0.044 | boundary | MID|MutS; MID|ABC_MSH5_euk; | 0.0 |
| 66 | fp_D | O43196 | 639 | ESISLGLSTFMIDLNQ | CCCCCCCCHHHHHHHH | c1a-AT-4 | multi | 0.383 | 0.073 | 0.05 | boundary | MID|MutS; MID|ABC_MSH5_euk; | 0.0 |
| 67 | fp_D | O43196 | 641 | ISLGLSTFMIDLNQ | CCCCCCHHHHHHHH | c2-AT-4 | multi | 0.396 | 0.065 | 0.053 | boundary | MID|MutS; MID|ABC_MSH5_euk; | 0.0 |
| 68 | fp_D | O43196 | 641 | ISLGLSTFMIDLNQVA | CCCCCCHHHHHHHHHH | c1aR-5 | multi-selected | 0.361 | 0.059 | 0.064 | boundary | MID|MutS; MID|ABC_MSH5_euk; | 0.0 |
| 69 | fp_D | O43196 | 644 | GLSTFMIDLNQVAK | CCCHHHHHHHHHHH | c3-4 | multi-selected | 0.341 | 0.037 | 0.074 | __ | MID|MutS; MID|ABC_MSH5_euk; | 0.0 |
| 70 | fp_D | O43196 | 648 | FMIDLNQVAKAVNNAT | HHHHHHHHHHHHHCCC | c1aR-4 | multi-selected | 0.225 | 0.018 | 0.086 | boundary | MID|MutS; MID|ABC_MSH5_euk; | 0.0 |
| 71 | fp_D | O43196 | 648 | FMIDLNQVAKAVNN | HHHHHHHHHHHHHC | c3-AT | multi | 0.251 | 0.017 | 0.082 | boundary | MID|MutS; MID|ABC_MSH5_euk; | 0.0 |
| 72 | fp_D | O43196 | 655 | VAKAVNNATAQSLVLID | HHHHHHCCCCCCEEEEC | c1c-AT-4 | uniq | 0.079 | 0.015 | 0.131 | boundary | MID|MutS; MID|ABC_MSH5_euk; | 0.0 |
| 73 | fp_D | O43196 | 669 | LIDEFGKGTNTVDG | EECCCCCCCCCHHH | c3-AT | multi-selected | 0.058 | 0.023 | 0.119 | boundary | MID|MutS; MID|ABC_MSH5_euk; | 0.0 |
| 74 | fp_D | O43196 | 673 | FGKGTNTVDGLALLA | CCCCCCCHHHHHHHH | c1b-AT-4 | multi | 0.039 | 0.023 | 0.072 | boundary | MID|MutS; MID|ABC_MSH5_euk; | 0.0 |
| 75 | fp_O | O43196 | 679 | TVDGLALLAAVLRH | CHHHHHHHHHHHHH | c3-AT | uniq | 0.016 | 0.012 | 0.06 | ORD | MID|MutS; MID|ABC_MSH5_euk; | 0.0 |
| 76 | fp_beta_O | O43196 | 699 | TCPHIFVATNFLSLVQ | CCCCEEEECCHHHHHC | c1a-AT-4 | multi | 0.017 | 0.013 | 0.037 | ORD | MID|MutS; MID|ABC_MSH5_euk; | 0.571 |
| 77 | fp_beta_O | O43196 | 699 | TCPHIFVATNFLSLVQ | CCCCEEEECCHHHHHC | c1d-AT-4 | multi | 0.017 | 0.013 | 0.037 | ORD | MID|MutS; MID|ABC_MSH5_euk; | 0.571 |
| 78 | fp_O | O43196 | 700 | CPHIFVATNFLSLVQ | CCCEEEECCHHHHHC | c1b-AT-4 | multi | 0.018 | 0.012 | 0.037 | ORD | MID|MutS; MID|ABC_MSH5_euk; | 0.429 |
| 79 | fp_O | O43196 | 701 | PHIFVATNFLSLVQLQL | CCEEEECCHHHHHCCCC | c1c-4 | multi-selected | 0.025 | 0.02 | 0.038 | ORD | MID|MutS; MID|ABC_MSH5_euk; | 0.25 |
| 80 | fp_O | O43196 | 701 | PHIFVATNFLSLVQ | CCEEEECCHHHHHC | c2-AT-4 | multi | 0.019 | 0.012 | 0.037 | ORD | MID|MutS; MID|ABC_MSH5_euk; | 0.429 |
| 81 | fp_O | O43196 | 702 | HIFVATNFLSLVQLQL | CEEEECCHHHHHCCCC | c1a-AT-5 | multi | 0.026 | 0.02 | 0.036 | ORD | MID|MutS; MID|ABC_MSH5_euk; | 0.143 |
| 82 | fp_O | O43196 | 702 | HIFVATNFLSLVQLQL | CEEEECCHHHHHCCCC | c1d-AT-5 | multi | 0.026 | 0.02 | 0.036 | ORD | MID|MutS; MID|ABC_MSH5_euk; | 0.143 |
| 83 | fp_O | O43196 | 703 | IFVATNFLSLVQLQL | EEEECCHHHHHCCCC | c1b-AT-5 | multi | 0.027 | 0.021 | 0.036 | ORD | MID|MutS; MID|ABC_MSH5_euk; | 0.0 |
| 84 | fp_O | O43196 | 705 | VATNFLSLVQLQLLP | EECCHHHHHCCCCCC | c1b-4 | multi | 0.031 | 0.03 | 0.035 | ORD | MID|MutS; MID|ABC_MSH5_euk; | 0.0 |
| 85 | fp_O | O43196 | 706 | ATNFLSLVQLQLLP | ECCHHHHHCCCCCC | c2-4 | multi-selected | 0.032 | 0.031 | 0.034 | ORD | MID|MutS; MID|ABC_MSH5_euk; | 0.0 |
| 86 | fp_beta_O | O43196 | 736 | NDLVFFYQVCEGVAKAS | CEEEEEEECCCCCCCCC | c1cR-4 | multi | 0.036 | 0.052 | 0.081 | ORD | MID|MutS; boundary|ABC_MSH5_euk; | 0.5 |
| 87 | fp_O | O43196 | 737 | DLVFFYQVCEGVAKAS | EEEEEEECCCCCCCCC | c1aR-4 | multi-selected | 0.038 | 0.053 | 0.083 | ORD | MID|MutS; boundary|ABC_MSH5_euk; | 0.429 |
| 88 | cand_D | O43196 | 798 | NCQTLVDKFMKLDLED *+++*++*+* | HHHHHHHHHHCCCCCC | c1a-4 | multi-selected | 0.351 | 0.114 | 0.212 | DISO | boundary|MutS; boundary|ABC_MSH5_euk; | 0.0 |
| 89 | cand_D | O43196 | 799 | CQTLVDKFMKLDLED *+++*++*+* | HHHHHHHHHCCCCCC | c1b-4 | multi | 0.336 | 0.114 | 0.21 | DISO | boundary|MutS; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment