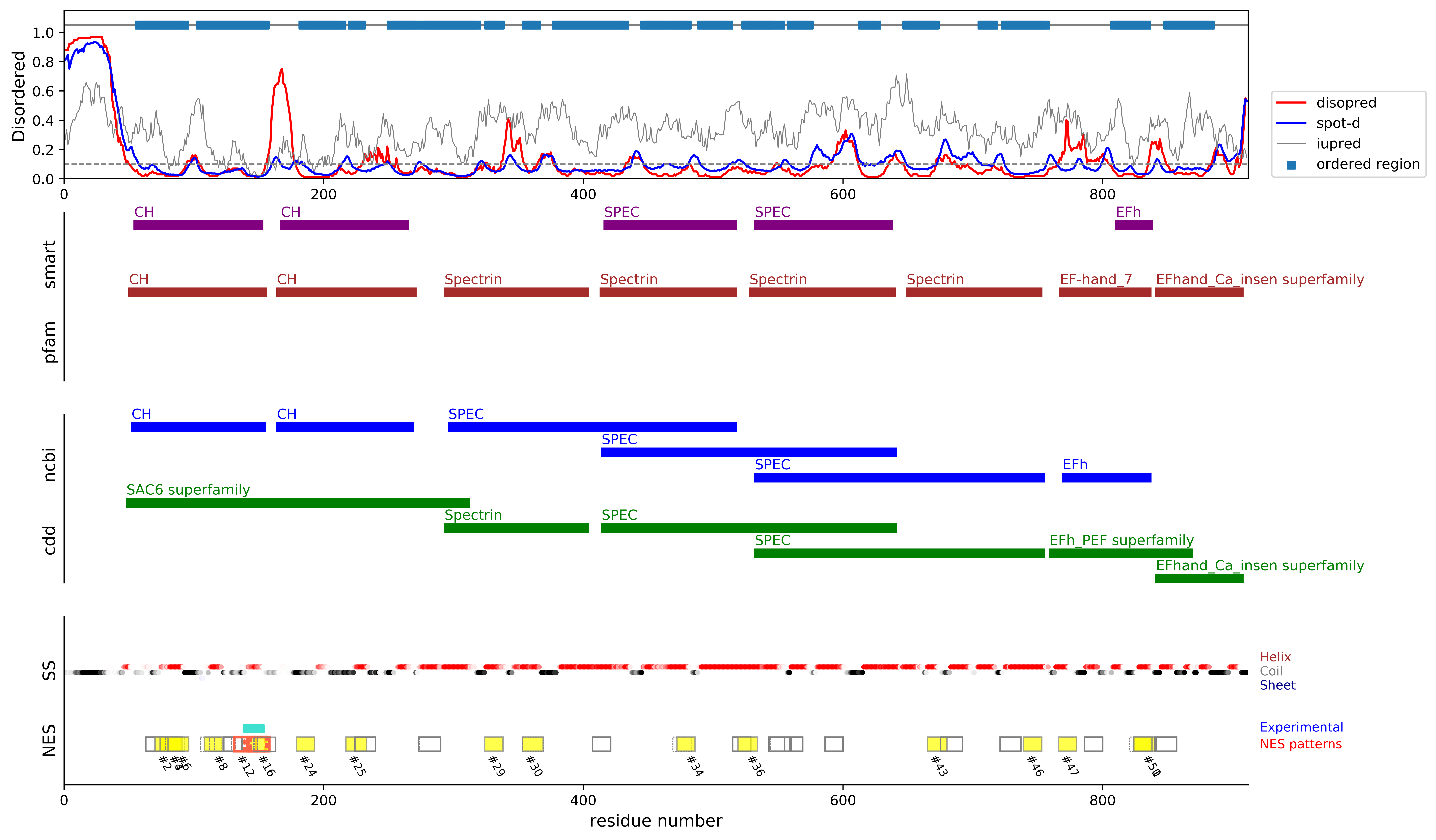

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | O43707 | 63 | HLRKAGTQIENIDE | HHHHCCCCCCCHHH | c3-AT | uniq | 0.034 | 0.072 | 0.329 | boundary | boundary|SAC6 superfamily; | 0.0 |
| 2 | fp_D | O43707 | 70 | QIENIDEDFRDGLKLML | CCCCHHHHHHHHHHHHH | c1c-5 | multi-selected | 0.026 | 0.043 | 0.217 | boundary | MID|SAC6 superfamily; | 0.0 |
| 3 | fp_D | O43707 | 74 | IDEDFRDGLKLMLLLE | HHHHHHHHHHHHHHHH | c1a-4 | multi-selected | 0.024 | 0.032 | 0.153 | boundary | MID|SAC6 superfamily; | 0.0 |
| 4 | fp_D | O43707 | 74 | IDEDFRDGLKLMLLLEV | HHHHHHHHHHHHHHHHH | c1c-4 | multi-selected | 0.025 | 0.032 | 0.156 | boundary | MID|SAC6 superfamily; | 0.0 |
| 5 | fp_D | O43707 | 78 | FRDGLKLMLLLEVIS | HHHHHHHHHHHHHHH | c1b-4 | multi | 0.027 | 0.029 | 0.131 | boundary | MID|SAC6 superfamily; | 0.0 |
| 6 | fp_D | O43707 | 80 | DGLKLMLLLEVISGER | HHHHHHHHHHHHHCCC | c1aR-4 | multi-selected | 0.041 | 0.034 | 0.155 | boundary | MID|SAC6 superfamily; | 0.0 |
| 7 | fp_D | O43707 | 105 | RVHKINNVNKALDFIA | CEEHHHCHHHHHHHHH | c1d-5 | multi | 0.036 | 0.04 | 0.242 | boundary | MID|SAC6 superfamily; | 0.0 |
| 8 | fp_D | O43707 | 108 | KINNVNKALDFIAS | HHHCHHHHHHHHHH | c3-4 | multi-selected | 0.028 | 0.037 | 0.183 | boundary | MID|SAC6 superfamily; | 0.0 |
| 9 | fp_D | O43707 | 112 | VNKALDFIASKGVKLVS | HHHHHHHHHHCCCCCCC | c1c-AT-4 | multi | 0.036 | 0.04 | 0.129 | boundary | MID|SAC6 superfamily; | 0.0 |
| 10 | fp_D | O43707 | 116 | LDFIASKGVKLVSIGA | HHHHHHCCCCCCCCCC | c1a-AT-4 | multi | 0.046 | 0.044 | 0.131 | boundary | MID|SAC6 superfamily; | 0.0 |
| 11 | fp_O | O43707 | 123 | GVKLVSIGAEEIVD | CCCCCCCCCCCCCC | c3-AT | uniq | 0.061 | 0.056 | 0.161 | ORD | MID|SAC6 superfamily; | 0.0 |
| 12 | cand_D | O43707 | 131 | AEEIVDGNAKMTLGMIW +++++++ | CCCCCCCCCHHHHHHHH | c1c-AT-4 | uniq | 0.04 | 0.046 | 0.12 | boundary | MID|SAC6 superfamily; | 0.0 |
| 13 | cand_D | O43707 | 139 | AKMTLGMIWTIILRFA +++++++++++ | CHHHHHHHHHHHHHHC | c1aR-5 | multi | 0.024 | 0.024 | 0.047 | boundary | MID|SAC6 superfamily; | 0.0 |
| 14 | cand_D | O43707 | 141 | MTLGMIWTIILRFAI +++++++++++ | HHHHHHHHHHHHHCC | c1b-AT-4 | multi | 0.027 | 0.02 | 0.038 | boundary | MID|SAC6 superfamily; | 0.0 |
| 15 | cand_D | O43707 | 141 | MTLGMIWTIILRFAIQD +++++++++++ | HHHHHHHHHHHHHCCCC | c1c-4 | multi | 0.039 | 0.023 | 0.042 | boundary | MID|SAC6 superfamily; | 0.0 |
| 16 | cand_D | O43707 | 142 | TLGMIWTIILRFAIQD ++++++++++ | HHHHHHHHHHHHCCCC | c1a-5 | multi-selected | 0.041 | 0.022 | 0.041 | boundary | MID|SAC6 superfamily; | 0.0 |
| 17 | cand_D | O43707 | 142 | TLGMIWTIILRFAIQD ++++++++++ | HHHHHHHHHHHHCCCC | c1d-5 | multi | 0.041 | 0.022 | 0.041 | boundary | MID|SAC6 superfamily; | 0.0 |
| 18 | cand_D | O43707 | 142 | TLGMIWTIILRFAI ++++++++++ | HHHHHHHHHHHHCC | c2-AT-5 | multi | 0.028 | 0.02 | 0.036 | boundary | MID|SAC6 superfamily; | 0.0 |
| 19 | cand_D | O43707 | 144 | GMIWTIILRFAIQD ++++++++ | HHHHHHHHHHCCCC | c2-AT-5 | multi | 0.044 | 0.022 | 0.038 | boundary | MID|SAC6 superfamily; | 0.0 |
| 20 | fp_D | O43707 | 146 | IWTIILRFAIQDISVEE ++++++ | HHHHHHHHCCCCCCCCC | c1c-AT-4 | multi | 0.154 | 0.044 | 0.059 | boundary | MID|SAC6 superfamily; | 0.0 |
| 21 | fp_D | O43707 | 147 | WTIILRFAIQDISVEE +++++ | HHHHHHHCCCCCCCCC | c1a-4 | multi | 0.163 | 0.046 | 0.06 | boundary | MID|SAC6 superfamily; | 0.0 |
| 22 | fp_D | O43707 | 147 | WTIILRFAIQDISVEE +++++ | HHHHHHHCCCCCCCCC | c1d-AT-4 | multi | 0.163 | 0.046 | 0.06 | boundary | MID|SAC6 superfamily; | 0.0 |
| 23 | fp_D | O43707 | 149 | IILRFAIQDISVEE +++ | HHHHHCCCCCCCCC | c2-5 | multi | 0.184 | 0.05 | 0.064 | boundary | MID|SAC6 superfamily; | 0.0 |
| 24 | fp_D | O43707 | 179 | PYKNVNVQNFHISW | CCCCCCCCCCCCCC | c2-4 | uniq | 0.024 | 0.086 | 0.158 | boundary | MID|SAC6 superfamily; | 0.0 |
| 25 | fp_D | O43707 | 217 | KDDPVTNLNNAFEVAE | CCCCCCCHHHHHHHHH | c1d-4 | uniq | 0.067 | 0.086 | 0.254 | boundary | MID|SAC6 superfamily; | 0.0 |
| 26 | fp_D | O43707 | 224 | LNNAFEVAEKYLDIPK | HHHHHHHHHHHCCCCC | c1d-AT-4 | uniq | 0.116 | 0.059 | 0.197 | boundary | MID|SAC6 superfamily; | 0.0 |
| 27 | fp_D | O43707 | 273 | KAETAANRICKVLAVNQ | HHHHHHHHHHHHHHHHH | c1c-AT-4 | multi-selected | 0.059 | 0.075 | 0.311 | boundary | MID|SAC6 superfamily; boundary|Spectrin; | 0.0 |
| 28 | fp_D | O43707 | 274 | AETAANRICKVLAVNQ | HHHHHHHHHHHHHHHH | c1d-AT-4 | multi | 0.058 | 0.073 | 0.316 | boundary | MID|SAC6 superfamily; boundary|Spectrin; | 0.0 |
| 29 | fp_D | O43707 | 324 | TIQEMQQKLEDFRD | CHHHHHHHHHHHHH | c3-4 | uniq | 0.079 | 0.062 | 0.439 | boundary | boundary|SAC6 superfamily; MID|Spectrin; | 0.0 |
| 30 | fp_D | O43707 | 353 | LEINFNTLQTKLRLSN | HHHHHHHHHHHHHHCC | c1aR-4 | multi-selected | 0.093 | 0.064 | 0.337 | boundary | MID|Spectrin; | 0.0 |
| 31 | fp_D | O43707 | 353 | LEINFNTLQTKLRLSN | HHHHHHHHHHHHHHCC | c1d-4 | multi | 0.093 | 0.064 | 0.337 | boundary | MID|Spectrin; | 0.0 |
| 32 | fp_D | O43707 | 407 | RLERLDHLAEKFRQ | HHHHHHHHHHHHHH | c3-AT | uniq | 0.036 | 0.057 | 0.274 | boundary | boundary|Spectrin; boundary|SPEC; | 0.0 |
| 33 | fp_D | O43707 | 469 | RVEQIAAIAQELNE | HHHHHHHHHHHHHH | c3-AT | multi | 0.052 | 0.085 | 0.33 | boundary | MID|SPEC; | 0.0 |
| 34 | fp_D | O43707 | 472 | QIAAIAQELNELDY | HHHHHHHHHHHCCC | c3-4 | multi-selected | 0.062 | 0.1 | 0.314 | boundary | MID|SPEC; | 0.0 |
| 35 | fp_D | O43707 | 515 | ALEKTEKQLEAIDQ | HHHHHHHHHHHHHH | c3-AT | multi-selected | 0.064 | 0.108 | 0.444 | boundary | MID|SPEC; boundary|SPEC; | 0.0 |
| 36 | fp_D | O43707 | 519 | TEKQLEAIDQLHLEY | HHHHHHHHHHHHHHH | c1b-4 | multi-selected | 0.055 | 0.093 | 0.384 | boundary | MID|SPEC; boundary|SPEC; | 0.0 |
| 37 | fp_D | O43707 | 543 | WMESAMEDLQDMFIVHT | HHHHHHHHHHHHHHCCC | c1c-AT-5 | multi-selected | 0.071 | 0.087 | 0.308 | boundary | MID|SPEC; boundary|SPEC; | 0.0 |
| 38 | fp_D | O43707 | 543 | WMESAMEDLQDMFIVH | HHHHHHHHHHHHHHCC | c1a-AT-5 | multi-selected | 0.073 | 0.086 | 0.312 | boundary | MID|SPEC; boundary|SPEC; | 0.0 |
| 39 | fp_D | O43707 | 544 | MESAMEDLQDMFIVH | HHHHHHHHHHHHHCC | c1b-4 | multi | 0.074 | 0.089 | 0.307 | boundary | MID|SPEC; boundary|SPEC; | 0.0 |
| 40 | fp_D | O43707 | 544 | MESAMEDLQDMFIVHT | HHHHHHHHHHHHHCCC | c1d-4 | multi | 0.073 | 0.089 | 0.302 | boundary | MID|SPEC; boundary|SPEC; | 0.0 |
| 41 | fp_D | O43707 | 555 | FIVHTIEEIEGLIS | HHCCCHHHHHHHHH | c3-AT | uniq | 0.037 | 0.086 | 0.271 | boundary | MID|SPEC; MID|SPEC; | 0.0 |
| 42 | fp_D | O43707 | 586 | AILAIHKEAQRIAE | HHHHHHHHHHHHHH | c3-AT | uniq | 0.156 | 0.158 | 0.435 | boundary | MID|SPEC; MID|SPEC; | 0.0 |
| 43 | fp_D | O43707 | 665 | IQTKMEEIGRISIEM | HHHHHHHHHHHCCCC | c1b-4 | uniq | 0.092 | 0.139 | 0.375 | boundary | boundary|SPEC; MID|SPEC; | 0.0 |
| 44 | fp_D | O43707 | 675 | ISIEMNGTLEDQLSHLK | HCCCCCCCHHHHHHHHH | c1cR-5 | uniq | 0.117 | 0.189 | 0.405 | boundary | boundary|SPEC; MID|SPEC; | 0.0 |
| 45 | fp_D | O43707 | 721 | DNKHTNYTMEHIRVGW | CCCHHHHHHHHHHHHH | c1a-AT-4 | uniq | 0.029 | 0.071 | 0.283 | boundary | boundary|SPEC; | 0.0 |
| 46 | fp_D | O43707 | 739 | LLTTIARTINEVEN | HHHHHHHHHHHHHH | c3-4 | uniq | 0.019 | 0.041 | 0.278 | boundary | boundary|SPEC; boundary|EFh_PEF superfamily; | 0.0 |
| 47 | fp_D | O43707 | 766 | QMQEFRASFNHFDK | HHHHHHHHHHHHCC | c3-4 | uniq | 0.213 | 0.067 | 0.43 | boundary | boundary|SPEC; boundary|EFh_PEF superfamily; | 0.0 |
| 48 | fp_D | O43707 | 786 | GPEEFKACLISLGY | CHHHHHHHHHHCCC | c2-AT-4 | uniq | 0.143 | 0.069 | 0.249 | boundary | boundary|SPEC; MID|EFh_PEF superfamily; | 0.0 |
| 49 | fp_D | O43707 | 821 | NHSGLVTFQAFIDFMS | CCCCCCCHHHHHHHHH | c1d-4 | multi | 0.037 | 0.045 | 0.214 | boundary | MID|EFh_PEF superfamily; boundary|EFhand_Ca_insen superfamily; | 0.0 |
| 50 | fp_D | O43707 | 824 | GLVTFQAFIDFMSRET | CCCCHHHHHHHHHHHH | c1aR-4 | multi-selected | 0.075 | 0.041 | 0.258 | boundary | MID|EFh_PEF superfamily; boundary|EFhand_Ca_insen superfamily; | 0.0 |
| 51 | fp_D | O43707 | 824 | GLVTFQAFIDFMSR | CCCCHHHHHHHHHH | c3-4 | multi-selected | 0.051 | 0.038 | 0.242 | boundary | MID|EFh_PEF superfamily; boundary|EFhand_Ca_insen superfamily; | 0.0 |
| 52 | fp_D | O43707 | 840 | TDTDTADQVIASFKVLA | CCCCCHHHHHHHHHHHH | c1c-AT-4 | multi-selected | 0.141 | 0.073 | 0.318 | __ | boundary|EFh_PEF superfamily; boundary|EFhand_Ca_insen superfamily; | 0.0 |
| 53 | fp_D | O43707 | 841 | DTDTADQVIASFKVLA | CCCCHHHHHHHHHHHH | c1a-AT-4 | multi-selected | 0.134 | 0.072 | 0.317 | boundary | boundary|EFh_PEF superfamily; boundary|EFhand_Ca_insen superfamily; | 0.0 |
| 54 | fp_D | O43707 | 841 | DTDTADQVIASFKVLA | CCCCHHHHHHHHHHHH | c1d-AT-4 | multi | 0.134 | 0.072 | 0.317 | boundary | boundary|EFh_PEF superfamily; boundary|EFhand_Ca_insen superfamily; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment