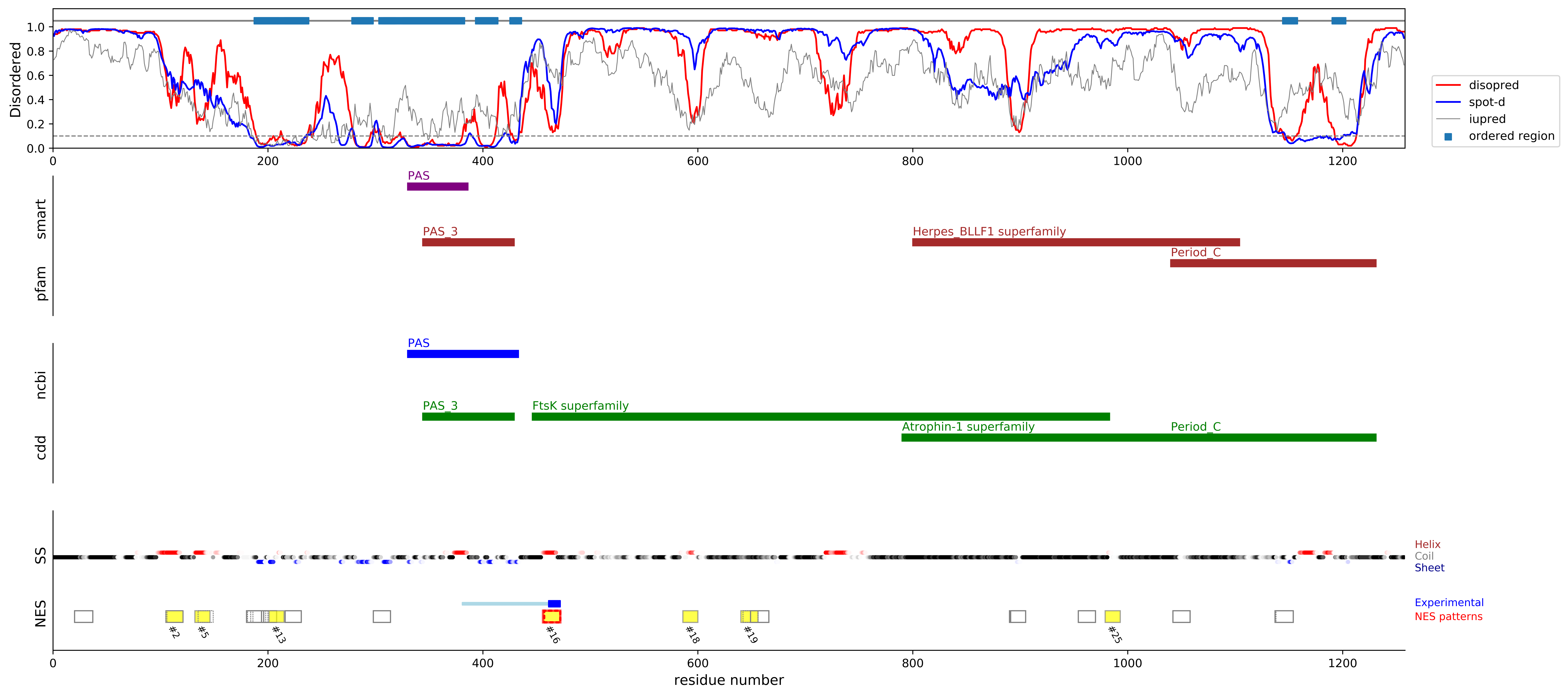

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | O54943 | 20 | APQPTQAVLQEDVDMSS | CCCCCCCCCCCCCCCCC | c1c-AT-4 | uniq | 0.972 | 0.969 | 0.891 | DISO | 0.0 | |
| 2 | fp_D | O54943 | 105 | AHKELIRTLKELKVHL | HHHHHHHHHHHHHHCC | c1a-4 | multi-selected | 0.46 | 0.553 | 0.458 | DISO | 0.0 | |
| 3 | fp_D | O54943 | 105 | AHKELIRTLKELKVHL | HHHHHHHHHHHHHHCC | c1d-AT-4 | multi | 0.46 | 0.553 | 0.458 | DISO | 0.0 | |
| 4 | fp_D | O54943 | 106 | HKELIRTLKELKVHL | HHHHHHHHHHHHHCC | c1b-4 | multi | 0.449 | 0.546 | 0.454 | DISO | 0.0 | |
| 5 | fp_D | O54943 | 132 | TLATLKYALRSVKQ | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.291 | 0.5 | 0.27 | DISO | 0.0 | |
| 6 | fp_D | O54943 | 135 | TLKYALRSVKQVKA | HHHHHHHHHHHHHC | c3-AT | multi | 0.331 | 0.471 | 0.236 | DISO | 0.0 | |
| 7 | fp_D | O54943 | 180 | TSEYIVKNADMFAVAV | CCCCCCCCCCEEEEEE | c1a-AT-4 | multi-selected | 0.18 | 0.065 | 0.102 | boundary | 0.143 | |
| 8 | fp_D | O54943 | 181 | SEYIVKNADMFAVAV | CCCCCCCCCEEEEEE | c1b-AT-4 | multi | 0.17 | 0.058 | 0.092 | boundary | 0.286 | |
| 9 | fp_beta_D | O54943 | 184 | IVKNADMFAVAVSL | CCCCCCEEEEEEEE | c2-AT-5 | multi | 0.107 | 0.036 | 0.067 | boundary | 0.571 | |
| 10 | fp_beta_D | O54943 | 184 | IVKNADMFAVAVSLVS | CCCCCCEEEEEEEECC | c1d-AT-5 | multi | 0.098 | 0.034 | 0.063 | boundary | 0.714 | |
| 11 | fp_beta_D | O54943 | 186 | KNADMFAVAVSLVS | CCCCEEEEEEEECC | c2-AT-4 | multi | 0.074 | 0.027 | 0.053 | boundary | 0.857 | |
| 12 | fp_beta_D | O54943 | 194 | AVSLVSGKILYISN | EEEECCCEEEEECC | c3-4 | uniq | 0.056 | 0.019 | 0.03 | boundary | 0.571 | |
| 13 | fp_D | O54943 | 201 | KILYISNQVASIFH | EEEEECCCHHHHCC | c3-4 | uniq | 0.085 | 0.025 | 0.035 | boundary | 0.286 | |
| 14 | fp_D | O54943 | 216 | KKDAFSDAKFVEFLA | CCCCCCCCCEEEEEC | c1b-AT-4 | uniq | 0.039 | 0.04 | 0.054 | boundary | 0.286 | |
| 15 | fp_D | O54943 | 298 | EQQGAESQLCCLLLAE | CCCCCCCCEEEEEEEE | c1a-AT-4 | uniq | 0.096 | 0.078 | 0.115 | boundary | 0.429 | |
| 16 | cand_D | O54943 | 456 | SVQELTEQIHRLLMQP ........*..*.* | HHHHHHHHHHHHHCCC | c1a-5 | multi-selected | 0.238 | 0.471 | 0.654 | DISO | boundary|PAS_3; boundary|FtsK superfamily; | 0.0 |
| 17 | cand_D | O54943 | 457 | VQELTEQIHRLLMQP .......*..*.* | HHHHHHHHHHHHCCC | c1b-AT-4 | multi | 0.219 | 0.447 | 0.643 | DISO | boundary|PAS_3; boundary|FtsK superfamily; | 0.0 |
| 18 | fp_D | O54943 | 586 | QISCLDSVIRYLES | HHCCHHHHHHHHHH | c3-4 | uniq | 0.442 | 0.843 | 0.231 | DISO | MID|FtsK superfamily; | 0.0 |
| 19 | fp_D | O54943 | 640 | SHTEVSAHLSSLTLPG | CCCCCCCCCCCCCCCC | c1a-4 | multi-selected | 0.957 | 0.974 | 0.509 | DISO | MID|FtsK superfamily; | 0.0 |
| 20 | fp_D | O54943 | 642 | TEVSAHLSSLTLPG | CCCCCCCCCCCCCC | c2-AT-4 | multi | 0.954 | 0.973 | 0.493 | DISO | MID|FtsK superfamily; | 0.0 |
| 21 | fp_D | O54943 | 649 | SSLTLPGKAESVVSLTS | CCCCCCCCCCCCCCCCC | c1c-AT-4 | uniq | 0.934 | 0.97 | 0.384 | DISO | MID|FtsK superfamily; | 0.0 |
| 22 | fp_beta_D | O54943 | 890 | FAAPLAPVMAFMLPS | CCCCCCCEEEECCCC | c1b-4 | multi-selected | 0.225 | 0.5 | 0.246 | DISO | MID|FtsK superfamily; MID|Atrophin-1 superfamily; | 0.571 |
| 23 | fp_beta_D | O54943 | 891 | AAPLAPVMAFMLPS | CCCCCCEEEECCCC | c2-AT-4 | multi-selected | 0.209 | 0.5 | 0.241 | DISO | MID|FtsK superfamily; MID|Atrophin-1 superfamily; | 0.571 |
| 24 | fp_D | O54943 | 954 | CACPVTPPAGTVALGR | CCCCCCCCCCCCCCCC | c1a-AT-4 | uniq | 0.981 | 0.914 | 0.558 | DISO | boundary|FtsK superfamily; MID|Atrophin-1 superfamily; | 0.0 |
| 25 | fp_D | O54943 | 979 | GSSPLQLNLLQLEE | CCCHHHHCCCCCCC | c2-4 | uniq | 0.968 | 0.877 | 0.627 | DISO | boundary|FtsK superfamily; MID|Atrophin-1 superfamily; | 0.0 |
| 26 | fp_D | O54943 | 1042 | DAISTSSDLLNLLLGE | CCCCCCCCCCCCCCCC | c1a-AT-4 | uniq | 0.89 | 0.856 | 0.438 | DISO | boundary|Atrophin-1 superfamily; boundary|Period_C; | 0.0 |
| 27 | fp_D | O54943 | 1137 | PIWLLMANTDDSIMMTY | CCEEECCCCCCCCEEEE | c1c-AT-5 | multi-selected | 0.112 | 0.098 | 0.316 | boundary | MID|Period_C; | 0.125 |
| 28 | fp_D | O54943 | 1138 | IWLLMANTDDSIMMTY | CEEECCCCCCCCEEEE | c1d-AT-4 | multi | 0.106 | 0.096 | 0.322 | boundary | MID|Period_C; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment