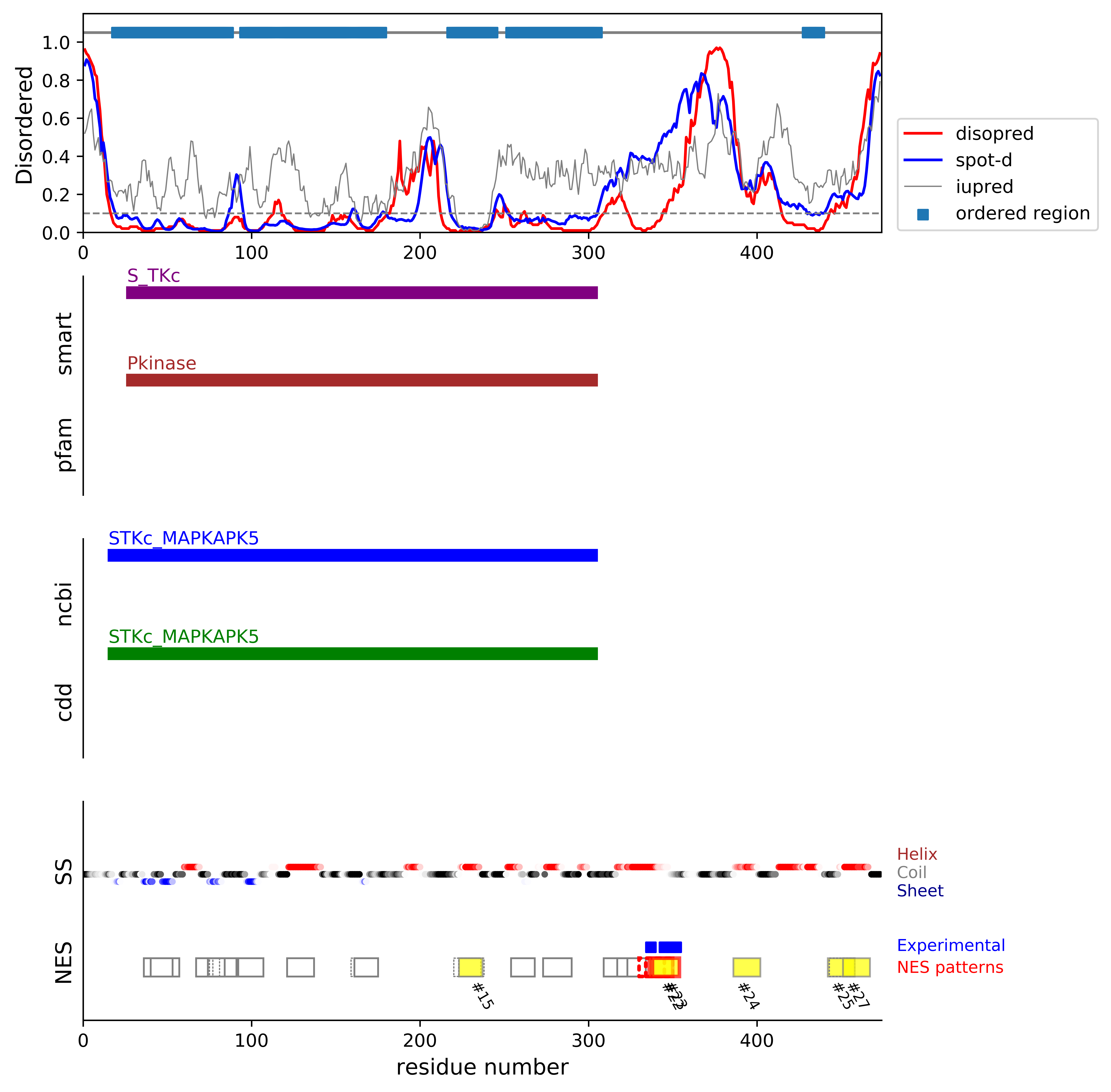

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_O | O54992 | 36 | VRVCVKKSTQERFALKI | EEEEEECCCCCEEEEEE | c1c-AT-4 | multi-selected | 0.017 | 0.031 | 0.252 | ORD | MID|STKc_MAPKAPK5; | 0.375 |
| 2 | fp_beta_O | O54992 | 40 | VKKSTQERFALKILLDR | EECCCCCEEEEEEECCC | c1c-AT-4 | multi-selected | 0.024 | 0.035 | 0.24 | ORD | MID|STKc_MAPKAPK5; | 0.625 |
| 3 | fp_D | O54992 | 67 | MMCATHPNIVQIIE | HHHCCCCCEEEEEE | c3-AT | multi | 0.011 | 0.013 | 0.168 | boundary | MID|STKc_MAPKAPK5; | 0.286 |
| 4 | fp_beta_D | O54992 | 67 | MMCATHPNIVQIIEVFA | HHHCCCCCEEEEEEEEC | c1c-AT-5 | multi-selected | 0.012 | 0.013 | 0.175 | boundary | MID|STKc_MAPKAPK5; | 0.5 |
| 5 | fp_beta_D | O54992 | 74 | NIVQIIEVFANSVQFPH | CEEEEEEEECCCCCCCC | c1c-5 | multi-selected | 0.026 | 0.058 | 0.183 | boundary | MID|STKc_MAPKAPK5; | 0.625 |
| 6 | fp_beta_D | O54992 | 75 | IVQIIEVFANSVQFPH | EEEEEEEECCCCCCCC | c1a-AT-5 | multi | 0.028 | 0.061 | 0.187 | boundary | MID|STKc_MAPKAPK5; | 0.571 |
| 7 | fp_beta_D | O54992 | 75 | IVQIIEVFANSVQFPH | EEEEEEEECCCCCCCC | c1d-5 | multi | 0.028 | 0.061 | 0.187 | boundary | MID|STKc_MAPKAPK5; | 0.571 |
| 8 | fp_D | O54992 | 77 | QIIEVFANSVQFPH | EEEEEECCCCCCCC | c2-AT-5 | multi | 0.03 | 0.068 | 0.196 | boundary | MID|STKc_MAPKAPK5; | 0.429 |
| 9 | fp_beta_D | O54992 | 92 | SSPRARLLIVMEMME | CCCCCEEEEEEEECC | c1b-AT-4 | uniq | 0.026 | 0.066 | 0.258 | boundary | MID|STKc_MAPKAPK5; | 0.857 |
| 10 | fp_O | O54992 | 121 | TEKQASQVTKQIALAL | CHHHHHHHHHHHHHHH | c1d-AT-4 | uniq | 0.021 | 0.025 | 0.276 | ORD | MID|STKc_MAPKAPK5; | 0.0 |
| 11 | fp_beta_D | O54992 | 159 | NSLDAPVKLCDFGFAK | CCCCCCEEEECCCCCC | c1a-AT-4 | multi | 0.024 | 0.06 | 0.133 | boundary | MID|STKc_MAPKAPK5; | 0.571 |
| 12 | fp_beta_D | O54992 | 161 | LDAPVKLCDFGFAK | CCCCEEEECCCCCC | c2-4 | multi-selected | 0.019 | 0.052 | 0.127 | boundary | MID|STKc_MAPKAPK5; | 0.571 |
| 13 | fp_D | O54992 | 220 | KSCDLWSLGVIIYVML | CCCCHHHHHHHHHHHH | c1d-4 | multi | 0.009 | 0.029 | 0.035 | boundary | MID|STKc_MAPKAPK5; | 0.0 |
| 14 | fp_D | O54992 | 223 | DLWSLGVIIYVMLCG | CHHHHHHHHHHHHCC | c1b-5 | multi | 0.009 | 0.022 | 0.017 | boundary | MID|STKc_MAPKAPK5; | 0.0 |
| 15 | fp_D | O54992 | 223 | DLWSLGVIIYVMLC | CHHHHHHHHHHHHC | c3-4 | multi-selected | 0.009 | 0.023 | 0.017 | boundary | MID|STKc_MAPKAPK5; | 0.0 |
| 16 | fp_D | O54992 | 254 | MRKKIMTGSFEFPE | HHHHHHCCEEECCC | c2-AT-4 | uniq | 0.059 | 0.065 | 0.356 | boundary | MID|STKc_MAPKAPK5; | 0.286 |
| 17 | fp_D | O54992 | 273 | ISEMAKDVVRKLLKVKP | HCHHHHHHHHHHCCCCC | c1c-AT-4 | uniq | 0.028 | 0.059 | 0.297 | boundary | boundary|STKc_MAPKAPK5; | 0.0 |
| 18 | fp_D | O54992 | 309 | ALDNVLPSAQLMMD | CCCCCCCCHHHHHH | c3-AT | uniq | 0.156 | 0.274 | 0.3 | boundary | boundary|STKc_MAPKAPK5; | 0.0 |
| 19 | fp_D | O54992 | 317 | AQLMMDKAVVAGIQQAH | HHHHHHHHHHHHHHHHH | c1cR-4 | uniq | 0.087 | 0.36 | 0.309 | boundary | boundary|STKc_MAPKAPK5; | 0.0 |
| 20 | cand_D | O54992 | 330 | QQAHAEQLANMRIQD * | HHHHHHHHHHHHHHH | c1b-AT-4 | multi | 0.035 | 0.408 | 0.358 | DISO | boundary|STKc_MAPKAPK5; | 0.0 |
| 21 | cand_D | O54992 | 334 | AEQLANMRIQDLKVSL * * * | HHHHHHHHHHHHHHCC | c1a-AT-4 | multi | 0.074 | 0.448 | 0.356 | DISO | boundary|STKc_MAPKAPK5; | 0.0 |
| 22 | cand_D | O54992 | 336 | QLANMRIQDLKVSL * * * | HHHHHHHHHHHHCC | c2-5 | multi-selected | 0.083 | 0.457 | 0.36 | DISO | boundary|STKc_MAPKAPK5; | 0.0 |
| 23 | cand_D | O54992 | 338 | ANMRIQDLKVSLKPLH * * * | HHHHHHHHHHCCCCCC | c1aR-5 | multi-selected | 0.122 | 0.495 | 0.348 | DISO | boundary|STKc_MAPKAPK5; | 0.0 |
| 24 | fp_D | O54992 | 386 | SNVALEKLRDVIAQCI | CHHHHHHHHHHHHHHC | c1aR-4 | uniq | 0.301 | 0.279 | 0.337 | DISO | 0.0 | |
| 25 | fp_D | O54992 | 442 | NGRGFTDKVDRLKLAE | CCCCCCCHHHHHHHHH | c1a-4 | multi-selected | 0.147 | 0.19 | 0.29 | boundary | 0.0 | |
| 26 | fp_D | O54992 | 443 | GRGFTDKVDRLKLAE | CCCCCCHHHHHHHHH | c1b-AT-4 | multi | 0.153 | 0.194 | 0.287 | boundary | 0.0 | |
| 27 | fp_D | O54992 | 451 | DRLKLAEVVKQVIEEQ | HHHHHHHHHHHHHHHH | c1aR-4 | uniq | 0.36 | 0.221 | 0.363 | boundary | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment