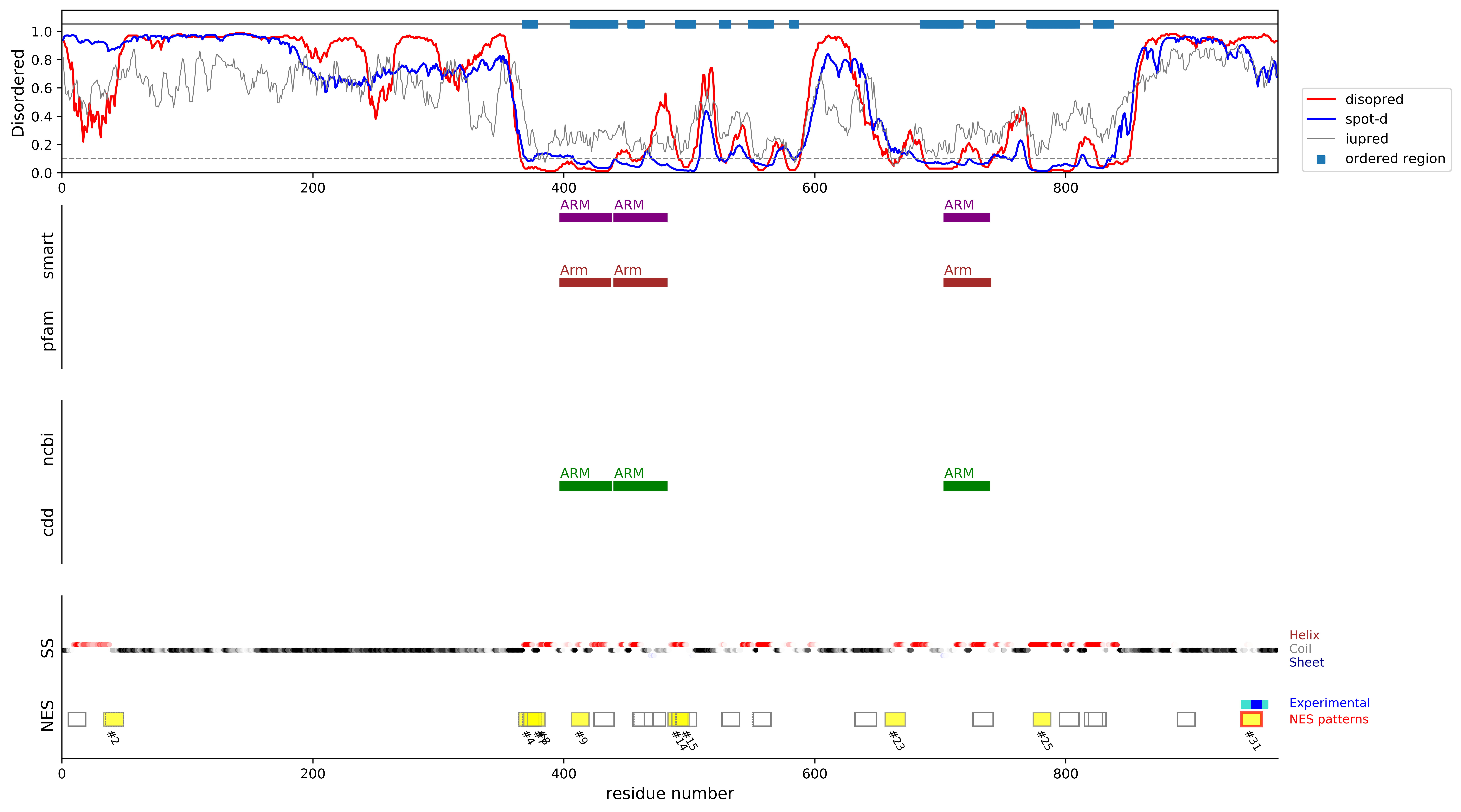

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | O60716 | 5 | EVESTASILASVKE | CCCCHHHHHHHHHH | c3-AT | uniq | 0.526 | 0.949 | 0.553 | DISO | 0.0 | |
| 2 | fp_D | O60716 | 33 | ERRHVSAQLERVRVSP | HHHHHHCCCCCCCCCC | c1a-4 | multi-selected | 0.572 | 0.888 | 0.65 | DISO | 0.0 | |
| 3 | fp_D | O60716 | 35 | RHVSAQLERVRVSP | HHHHCCCCCCCCCC | c2-AT-4 | multi | 0.599 | 0.884 | 0.662 | DISO | 0.0 | |
| 4 | fp_D | O60716 | 364 | RQPELPEVIAMLGFRL | CCCCHHHHHHHHCCCC | c1a-4 | multi-selected | 0.056 | 0.135 | 0.351 | boundary | 0.0 | |
| 5 | fp_D | O60716 | 364 | RQPELPEVIAMLGFRL | CCCCHHHHHHHHCCCC | c1d-4 | multi | 0.056 | 0.135 | 0.351 | boundary | 0.0 | |
| 6 | fp_D | O60716 | 367 | ELPEVIAMLGFRLDA | CHHHHHHHHCCCCHH | c1b-5 | multi | 0.037 | 0.111 | 0.262 | boundary | 0.0 | |
| 7 | fp_D | O60716 | 368 | LPEVIAMLGFRLDA | HHHHHHHHCCCCHH | c2-4 | multi-selected | 0.034 | 0.106 | 0.247 | boundary | 0.0 | |
| 8 | fp_D | O60716 | 371 | VIAMLGFRLDAVKS | HHHHHCCCCHHHHH | c3-4 | multi-selected | 0.031 | 0.112 | 0.189 | boundary | 0.0 | |
| 9 | fp_D | O60716 | 406 | KLKGIPVLVGLLDH | CCCCHHHHHHCCCC | c3-4 | uniq | 0.041 | 0.077 | 0.245 | boundary | small|ARM; | 0.0 |
| 10 | fp_D | O60716 | 424 | VHLGACGALKNISFGR | HHHHHHHHHHHHHCCC | c1a-AT-4 | uniq | 0.019 | 0.04 | 0.271 | boundary | 0.0 | |
| 11 | fp_D | O60716 | 455 | ALVRLLRKARDMDLTE | HHHHHHHHCCCCCCEE | c1a-AT-5 | multi-selected | 0.131 | 0.086 | 0.186 | boundary | small|ARM; | 0.0 |
| 12 | fp_D | O60716 | 456 | LVRLLRKARDMDLTE | HHHHHHHCCCCCCEE | c1b-AT-5 | multi | 0.135 | 0.089 | 0.188 | boundary | small|ARM; | 0.0 |
| 13 | fp_beta_D | O60716 | 464 | RDMDLTEVITGTLWNLS | CCCCCEEEECCCCCCCC | c1cR-5 | uniq | 0.318 | 0.093 | 0.201 | boundary | 0.5 | |
| 14 | fp_D | O60716 | 483 | DSIKMEIVDHALHALT | CCHHHHHHHHHHHHHH | c1aR-5 | multi-selected | 0.179 | 0.027 | 0.208 | boundary | 0.0 | |
| 15 | fp_D | O60716 | 486 | KMEIVDHALHALTD | HHHHHHHHHHHHHC | c3-4 | multi-selected | 0.121 | 0.021 | 0.206 | boundary | 0.0 | |
| 16 | fp_D | O60716 | 489 | IVDHALHALTDEVIIPH | HHHHHHHHHHCCCEECC | c1c-AT-5 | multi | 0.095 | 0.018 | 0.26 | boundary | 0.0 | |
| 17 | fp_D | O60716 | 490 | VDHALHALTDEVIIPH | HHHHHHHHHCCCEECC | c1a-AT-4 | multi | 0.089 | 0.017 | 0.261 | boundary | 0.0 | |
| 18 | fp_D | O60716 | 490 | VDHALHALTDEVIIPH | HHHHHHHHHCCCEECC | c1d-4 | multi | 0.089 | 0.017 | 0.261 | boundary | 0.0 | |
| 19 | fp_D | O60716 | 526 | VLTNTAGCLRNVSS | HHHCCCCCCCCCCC | c3-AT | uniq | 0.156 | 0.124 | 0.337 | boundary | 0.0 | |
| 20 | fp_D | O60716 | 550 | ECDGLVDALIFIVQA | HCCCHHHHHHHHHHH | c1b-AT-4 | multi | 0.044 | 0.062 | 0.158 | boundary | 0.0 | |
| 21 | fp_D | O60716 | 551 | CDGLVDALIFIVQA | CCCHHHHHHHHHHH | c2-AT-4 | multi-selected | 0.041 | 0.061 | 0.153 | boundary | 0.0 | |
| 22 | fp_D | O60716 | 632 | GKKPIEDPANDTVDFPK | CCCCCCCCCCCCCCCCC | c1c-AT-4 | uniq | 0.443 | 0.596 | 0.573 | DISO | 0.0 | |
| 23 | fp_D | O60716 | 656 | YELLFQPEVVRIYISL | CCCCCCCHHHHHHHHH | c1a-4 | uniq | 0.105 | 0.173 | 0.138 | __ | 0.0 | |
| 24 | fp_D | O60716 | 726 | VVKAASGALRNLAVDA | HHHHHHHHHHHHHCCC | c1a-AT-5 | uniq | 0.093 | 0.073 | 0.291 | boundary | 0.0 | |
| 25 | fp_D | O60716 | 774 | TVISILNTINEVIA | HHHHHHHHHHHHHH | c3-4 | uniq | 0.011 | 0.016 | 0.186 | boundary | 0.0 | |
| 26 | fp_D | O60716 | 795 | KLRETQGIEKLVLINK | HHHHCCCHHHHHHHHC | c1d-AT-5 | multi-selected | 0.039 | 0.054 | 0.389 | boundary | 0.0 | |
| 27 | fp_D | O60716 | 795 | KLRETQGIEKLVLIN | HHHHCCCHHHHHHHH | c1b-AT-5 | multi-selected | 0.031 | 0.054 | 0.385 | boundary | 0.0 | |
| 28 | fp_D | O60716 | 815 | SEKEVRAAALVLQT | CHHHHHHHHHHHHH | c2-AT-4 | multi-selected | 0.169 | 0.05 | 0.347 | boundary | 0.0 | |
| 29 | fp_D | O60716 | 818 | EVRAAALVLQTIWG | HHHHHHHHHHHHHC | c3-AT | multi-selected | 0.126 | 0.039 | 0.322 | boundary | 0.0 | |
| 30 | fp_D | O60716 | 889 | QMSNMGSNTKSLDN | CCCCCCCCCCCCCC | c3-AT | uniq | 0.949 | 0.953 | 0.802 | DISO | 0.0 | |
| 31 | cand_D | O60716 | 940 | GQESLEEELDVLVLDD ++++++++*+*++ | CCCCHHHHCCCCCCCC | c1a-4 | uniq | 0.956 | 0.791 | 0.756 | DISO | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment