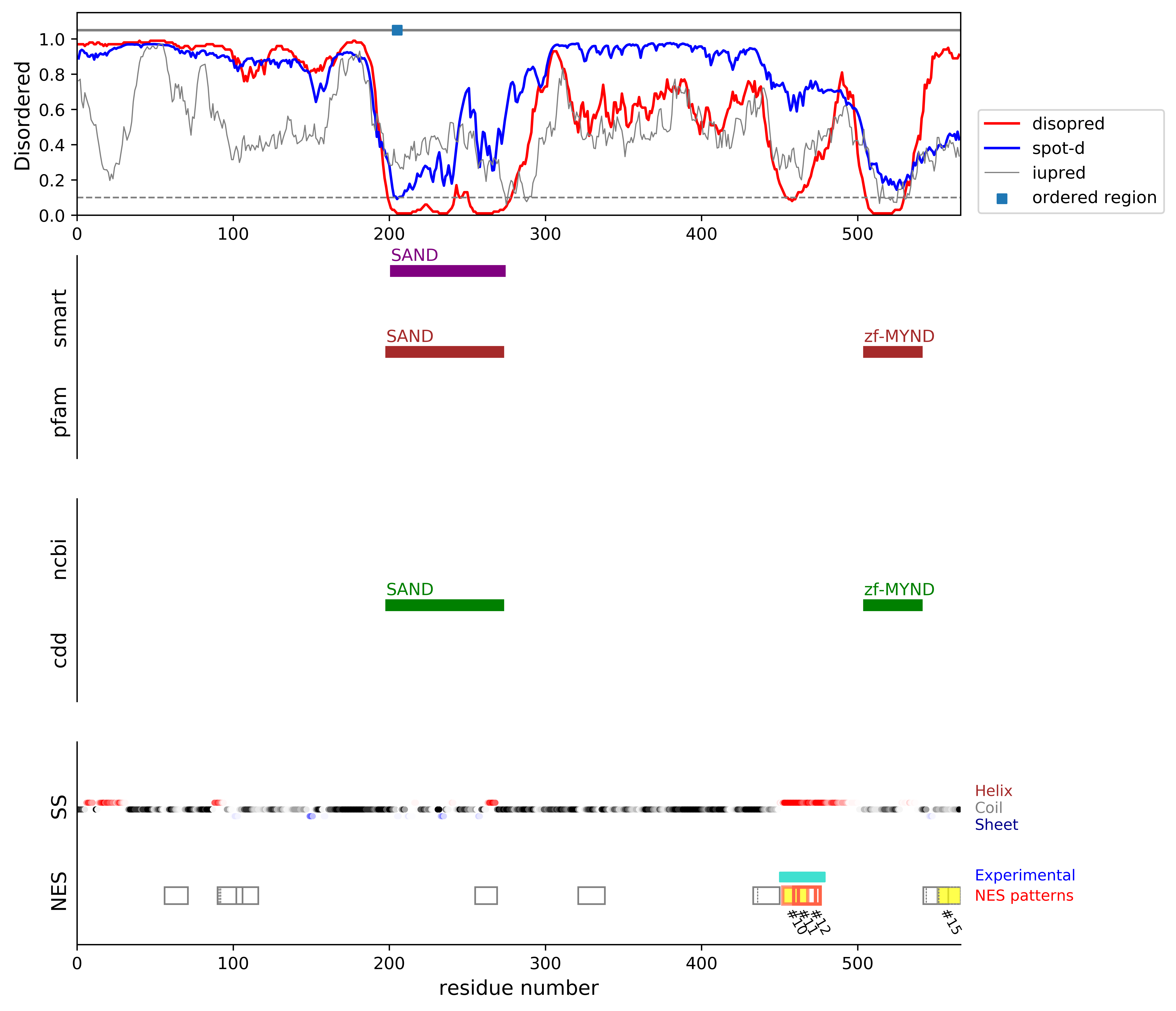

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | O75398 | 56 | AERETPRVTAVAVMA | CCCCCCCCCCCCCCC | c1b-AT-4 | uniq | 0.968 | 0.945 | 0.728 | DISO | 0.0 | |
| 2 | fp_D | O75398 | 90 | AAAAAFAEVTTVTVAN | HHHHHCCCCCEEEECC | c1a-AT-4 | multi-selected | 0.92 | 0.877 | 0.427 | DISO | 0.143 | |
| 3 | fp_D | O75398 | 91 | AAAAFAEVTTVTVAN | HHHHCCCCCEEEECC | c1b-4 | multi | 0.917 | 0.875 | 0.416 | DISO | 0.286 | |
| 4 | fp_D | O75398 | 92 | AAAFAEVTTVTVAN | HHHCCCCCEEEECC | c2-AT-4 | multi | 0.914 | 0.874 | 0.406 | DISO | 0.286 | |
| 5 | fp_D | O75398 | 102 | TVANVGAAADNVFT | EECCCCCCCCCCCC | c3-AT | uniq | 0.815 | 0.865 | 0.381 | DISO | 0.0 | |
| 6 | fp_D | O75398 | 255 | SIRYAGRPLQCLIQ | CCEECCCCHHHHHH | c3-AT | uniq | 0.012 | 0.375 | 0.298 | DISO | boundary|SAND; | 0.143 |
| 7 | fp_D | O75398 | 321 | NITLLPATAATTFTVTP | CCCCCCCCCCCCCCCCC | c1c-AT-5 | uniq | 0.576 | 0.931 | 0.463 | DISO | 0.0 | |
| 8 | fp_D | O75398 | 433 | PTKAAPPALVNGLELSE | CCCCCCCCCCCCCCCCC | c1c-AT-4 | multi-selected | 0.505 | 0.828 | 0.541 | DISO | 0.0 | |
| 9 | fp_D | O75398 | 436 | AAPPALVNGLELSE | CCCCCCCCCCCCCC | c2-AT-4 | multi | 0.461 | 0.804 | 0.526 | DISO | 0.0 | |
| 10 | cand_D | O75398 | 452 | SWLYLEEMVNSLLNTA +++++++++++++++ | HHHHHHHHHHHHHHHH | c1aR-4 | uniq | 0.117 | 0.67 | 0.308 | DISO | 0.0 | |
| 11 | cand_D | O75398 | 459 | MVNSLLNTAQQLKT ++++++++++++++ | HHHHHHHHHHHHHH | c3-AT | multi-selected | 0.164 | 0.689 | 0.333 | DISO | 0.0 | |
| 12 | cand_D | O75398 | 462 | SLLNTAQQLKTLFE ++++++++++++++ | HHHHHHHHHHHHHH | c3-AT | multi-selected | 0.214 | 0.71 | 0.352 | DISO | 0.0 | |
| 13 | fp_D | O75398 | 542 | QSAAVTVQADEVHVAE | CCCEEEECCCCCCCCC | c1a-AT-4 | multi-selected | 0.845 | 0.374 | 0.353 | DISO | 0.429 | |
| 14 | fp_D | O75398 | 544 | AAVTVQADEVHVAE | CEEEECCCCCCCCC | c2-AT-4 | multi | 0.871 | 0.38 | 0.357 | DISO | 0.286 | |
| 15 | fp_D | O75398 | 551 | DEVHVAESVMEKVTVXX | CCCCCCCCCCCCCCC | c1c-4-Ct | multi-selected | 0.915 | 0.424 | 0.391 | DISO | 0.0 | |
| 16 | fp_D | O75398 | 552 | EVHVAESVMEKVTVXX | CCCCCCCCCCCCCC | c1a-AT-5-Ct | multi | 0.915 | 0.428 | 0.387 | DISO | 0.0 | |
| 17 | fp_D | O75398 | 552 | EVHVAESVMEKVTVXX | CCCCCCCCCCCCCC | c1d-AT-5-Ct | multi | 0.915 | 0.428 | 0.387 | DISO | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment