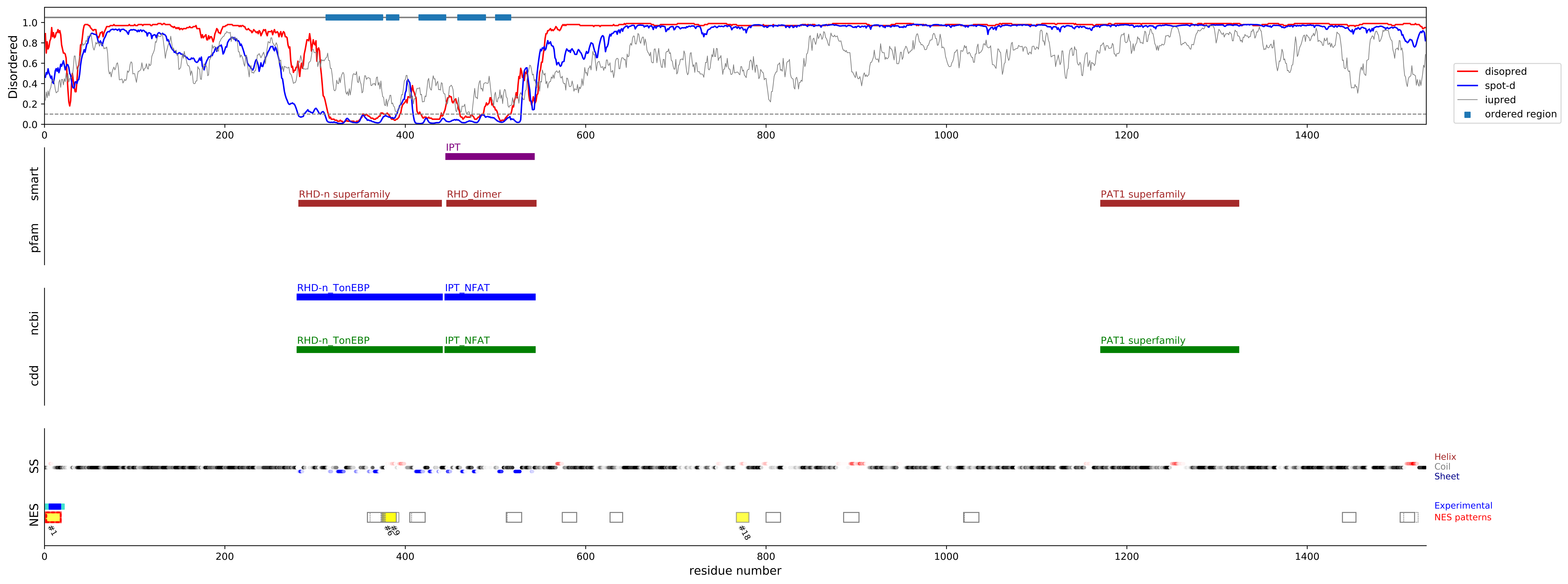

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | cand_D | O94916 | 1 | MPSDFISLLSADLDLES +++++++**+++*+*++ | CCCCCHHHCCCCCCCCC | c1c-4 | multi-selected | 0.855 | 0.493 | 0.35 | DISO | 0.0 | |
| 2 | cand_D | O94916 | 2 | PSDFISLLSADLDLES ++++++**+++*+*++ | CCCCHHHCCCCCCCCC | c1d-4 | multi | 0.853 | 0.495 | 0.357 | DISO | 0.0 | |
| 3 | fp_beta_D | O94916 | 358 | KEVDIEGTTVIEVGLDP | EEEEECCEEEEEEEECC | c1c-AT-4 | multi-selected | 0.072 | 0.027 | 0.41 | boundary | MID|RHD-n_TonEBP; | 0.75 |
| 4 | fp_beta_D | O94916 | 358 | KEVDIEGTTVIEVGL | EEEEECCEEEEEEEE | c1b-AT-4 | multi | 0.067 | 0.026 | 0.415 | boundary | MID|RHD-n_TonEBP; | 0.714 |
| 5 | fp_beta_D | O94916 | 361 | DIEGTTVIEVGLDP | EECCEEEEEEEECC | c2-AT-5 | multi | 0.069 | 0.024 | 0.401 | boundary | MID|RHD-n_TonEBP; | 0.857 |
| 6 | fp_D | O94916 | 374 | PSNNMTLAVDCVGILK | CCCCCHHHHHHHHHHH | c1a-4 | multi-selected | 0.081 | 0.069 | 0.224 | boundary | MID|RHD-n_TonEBP; | 0.0 |
| 7 | fp_D | O94916 | 374 | PSNNMTLAVDCVGILK | CCCCCHHHHHHHHHHH | c1d-AT-4 | multi | 0.081 | 0.069 | 0.224 | boundary | MID|RHD-n_TonEBP; | 0.0 |
| 8 | fp_D | O94916 | 375 | SNNMTLAVDCVGILK | CCCCHHHHHHHHHHH | c1b-AT-4 | multi | 0.079 | 0.07 | 0.217 | boundary | MID|RHD-n_TonEBP; | 0.0 |
| 9 | fp_D | O94916 | 376 | NNMTLAVDCVGILK | CCCHHHHHHHHHHH | c2-4 | multi-selected | 0.076 | 0.071 | 0.213 | boundary | MID|RHD-n_TonEBP; | 0.0 |
| 10 | fp_D | O94916 | 377 | NMTLAVDCVGILKLRN | CCHHHHHHHHHHHCCC | c1a-AT-5 | multi | 0.071 | 0.079 | 0.193 | boundary | MID|RHD-n_TonEBP; | 0.0 |
| 11 | fp_D | O94916 | 378 | MTLAVDCVGILKLRN | CHHHHHHHHHHHCCC | c1b-4 | multi | 0.069 | 0.079 | 0.193 | boundary | MID|RHD-n_TonEBP; | 0.0 |
| 12 | fp_beta_D | O94916 | 405 | KKKSTRARLVFRVNIMR | CCCCCCEEEEEEEEEEC | c1c-AT-4 | multi-selected | 0.193 | 0.083 | 0.281 | boundary | boundary|RHD-n_TonEBP; | 0.75 |
| 13 | fp_beta_D | O94916 | 407 | KSTRARLVFRVNIMR | CCCCEEEEEEEEEEC | c1b-AT-4 | multi | 0.183 | 0.044 | 0.281 | boundary | boundary|RHD-n_TonEBP; | 1.0 |
| 14 | fp_beta_D | O94916 | 512 | HDQHITLPVSVGIYVVT | CCCCCCCCEEEEEEEEC | c1c-4 | multi-selected | 0.231 | 0.043 | 0.235 | boundary | boundary|IPT_NFAT; | 0.5 |
| 15 | fp_beta_D | O94916 | 513 | DQHITLPVSVGIYVVT | CCCCCCCEEEEEEEEC | c1d-AT-4 | multi | 0.241 | 0.042 | 0.233 | boundary | boundary|IPT_NFAT; | 0.571 |
| 16 | fp_D | O94916 | 574 | AMKTTGCNLDKVNIIP | HCCCCCCCCCCCCCCC | c1a-AT-5 | uniq | 0.978 | 0.696 | 0.393 | DISO | boundary|IPT_NFAT; | 0.0 |
| 17 | fp_D | O94916 | 627 | SVGSTQQTLENISN | CCCCCCCCCCCCCC | c3-AT | uniq | 0.974 | 0.905 | 0.574 | DISO | 0.0 | |
| 18 | fp_D | O94916 | 767 | SVSQLQNTIQQLQA | CCHHHHHHHHHCCC | c3-4 | uniq | 0.971 | 0.94 | 0.513 | DISO | 0.0 | |
| 19 | fp_D | O94916 | 800 | QVLEAQQQLSSVLFSA | HHHHCCCCCCCCCCCC | c1a-AT-5 | uniq | 0.971 | 0.966 | 0.416 | DISO | 0.0 | |
| 20 | fp_D | O94916 | 886 | QQQVMESSAAMVMEMQQ | HHHHCCHHHHHHHHHHH | c1c-AT-4 | uniq | 0.99 | 0.963 | 0.601 | DISO | 0.0 | |
| 21 | fp_D | O94916 | 1019 | PQVNLFSSTKSMMSVQN | CCCCCCCCCCCCCCCCC | c1c-AT-4 | multi-selected | 0.974 | 0.968 | 0.704 | DISO | 0.0 | |
| 22 | fp_D | O94916 | 1020 | QVNLFSSTKSMMSVQN | CCCCCCCCCCCCCCCC | c1d-AT-5 | multi | 0.974 | 0.968 | 0.702 | DISO | 0.0 | |
| 23 | fp_D | O94916 | 1439 | SSQQTSGMFLFGIQN | CCCCCCCCCCCCCCC | c1b-AT-4 | uniq | 0.973 | 0.933 | 0.493 | DISO | 0.0 | |
| 24 | fp_D | O94916 | 1503 | SSINTNQNIEKIDLLV | CCCCCCHHHHHHHHHH | c1a-AT-4 | multi-selected | 0.985 | 0.838 | 0.544 | DISO | 0.0 | |
| 25 | fp_D | O94916 | 1507 | TNQNIEKIDLLVSLQN | CCHHHHHHHHHHHHHH | c1d-4 | multi | 0.986 | 0.81 | 0.468 | DISO | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment