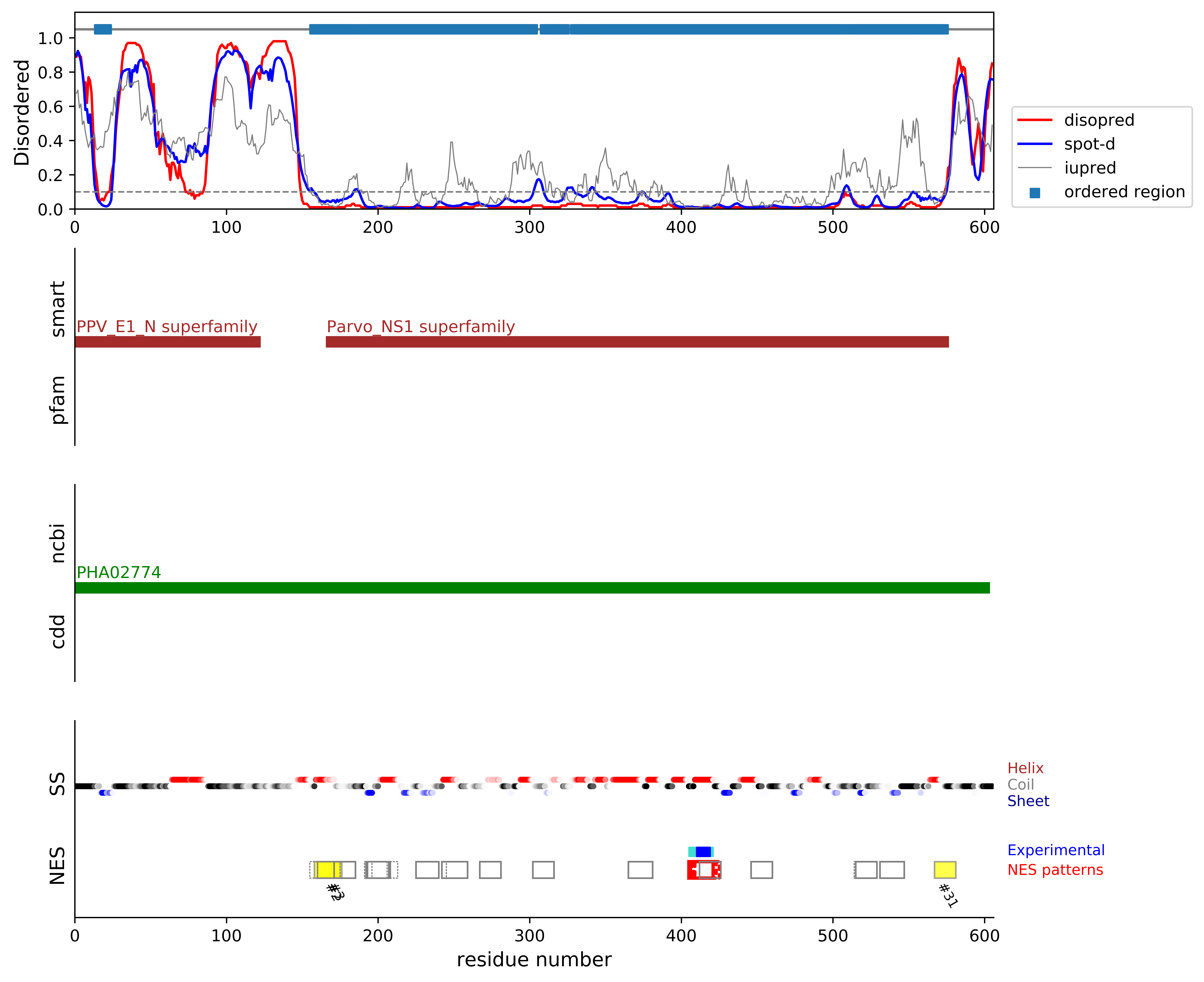

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | P03116 | 155 | KSKNATVFKLGLFK | HCCCHHHHHHHHHH | c2-AT-4 | multi | 0.01 | 0.069 | 0.054 | boundary | MID|PHA02774; | 0.0 |
| 2 | fp_D | P03116 | 158 | NATVFKLGLFKSLFLCS | CHHHHHHHHHHHHHCCC | c1c-4 | multi-selected | 0.01 | 0.053 | 0.033 | boundary | MID|PHA02774; | 0.0 |
| 3 | fp_D | P03116 | 160 | TVFKLGLFKSLFLCSF | HHHHHHHHHHHHCCCC | c1aR-4 | multi-selected | 0.01 | 0.048 | 0.027 | boundary | MID|PHA02774; | 0.0 |
| 4 | fp_D | P03116 | 160 | TVFKLGLFKSLFLCS | HHHHHHHHHHHHCCC | c1b-5 | multi | 0.01 | 0.048 | 0.027 | boundary | MID|PHA02774; | 0.0 |
| 5 | fp_D | P03116 | 171 | FLCSFHDITRLFKN | HCCCCCCCCCCCCC | c3-AT | uniq | 0.016 | 0.062 | 0.086 | boundary | MID|PHA02774; | 0.0 |
| 6 | fp_O | P03116 | 191 | QWVLAVFGLAEVFFEA | CCEEEECCCCHHHHHH | c1a-AT-4 | multi | 0.009 | 0.014 | 0.038 | ORD | MID|PHA02774; | 0.286 |
| 7 | fp_O | P03116 | 192 | WVLAVFGLAEVFFEAS | CEEEECCCCHHHHHHH | c1aR-4 | multi-selected | 0.009 | 0.013 | 0.033 | ORD | MID|PHA02774; | 0.143 |
| 8 | fp_O | P03116 | 192 | WVLAVFGLAEVFFEA | CEEEECCCCHHHHHH | c1b-5 | multi | 0.009 | 0.014 | 0.034 | ORD | MID|PHA02774; | 0.143 |
| 9 | fp_O | P03116 | 192 | WVLAVFGLAEVFFE | CEEEECCCCHHHHH | c3-AT | multi | 0.009 | 0.014 | 0.035 | ORD | MID|PHA02774; | 0.286 |
| 10 | fp_O | P03116 | 193 | VLAVFGLAEVFFEA | EEEECCCCHHHHHH | c2-5 | multi-selected | 0.009 | 0.013 | 0.029 | ORD | MID|PHA02774; | 0.143 |
| 11 | fp_O | P03116 | 196 | VFGLAEVFFEASFELLK | ECCCCHHHHHHHHHHHH | c1c-AT-5 | multi | 0.009 | 0.012 | 0.019 | ORD | MID|PHA02774; | 0.0 |
| 12 | fp_beta_O | P03116 | 225 | HEGGTCAVYLICFNT | CCCCEEEEEEEECCC | c1b-AT-4 | uniq | 0.01 | 0.013 | 0.069 | ORD | MID|PHA02774; | 1.0 |
| 13 | fp_O | P03116 | 242 | SRETVRNLMANTLNVRE | CHHHHHHHHHHCCCCCC | c1c-4 | multi-selected | 0.01 | 0.024 | 0.187 | ORD | MID|PHA02774; | 0.0 |
| 14 | fp_O | P03116 | 245 | TVRNLMANTLNVRE | HHHHHHHHCCCCCC | c2-AT-5 | multi | 0.01 | 0.022 | 0.212 | ORD | MID|PHA02774; | 0.0 |
| 15 | fp_O | P03116 | 245 | TVRNLMANTLNVRE | HHHHHHHHCCCCCC | c3-AT | multi | 0.01 | 0.022 | 0.212 | ORD | MID|PHA02774; | 0.0 |
| 16 | fp_O | P03116 | 267 | KIRGLSAALFWFKS | CCCCCHHHHHHHHH | c2-AT-5 | uniq | 0.015 | 0.022 | 0.04 | ORD | MID|PHA02774; | 0.0 |
| 17 | fp_D | P03116 | 302 | LNESLQTEKFDFGT | HCCCCCCCCEEEHH | c2-AT-4 | uniq | 0.015 | 0.102 | 0.192 | boundary | MID|PHA02774; | 0.143 |
| 18 | fp_O | P03116 | 365 | VRHYLRAETQALSMPA | HHHHHHHHHHCCCHHH | c1a-AT-4 | uniq | 0.019 | 0.064 | 0.106 | ORD | MID|PHA02774; | 0.0 |
| 19 | cand_O | P03116 | 405 | QNIELITFINALKLWL +++++*++*++ | CCCCHHHHHHHHHHHH | c1aR-4 | multi-selected | 0.01 | 0.009 | 0.013 | ORD | MID|PHA02774; | 0.0 |
| 20 | cand_O | P03116 | 405 | QNIELITFINALKLWL +++++*++*++ | CCCCHHHHHHHHHHHH | c1a-4 | multi-selected | 0.01 | 0.009 | 0.013 | ORD | MID|PHA02774; | 0.0 |
| 21 | cand_O | P03116 | 405 | QNIELITFINALKLWL +++++*++*++ | CCCCHHHHHHHHHHHH | c1d-4 | multi | 0.01 | 0.009 | 0.013 | ORD | MID|PHA02774; | 0.0 |
| 22 | cand_O | P03116 | 406 | NIELITFINALKLWL +++++*++*++ | CCCHHHHHHHHHHHH | c1b-5 | multi | 0.01 | 0.009 | 0.013 | ORD | MID|PHA02774; | 0.0 |
| 23 | cand_O | P03116 | 407 | IELITFINALKLWL +++++*++*++ | CCHHHHHHHHHHHH | c2-AT-4 | multi | 0.01 | 0.009 | 0.014 | ORD | MID|PHA02774; | 0.0 |
| 24 | cand_O | P03116 | 408 | ELITFINALKLWLKGIP +++++*++*++ | CHHHHHHHHHHHHHCCC | c1cR-5 | multi | 0.01 | 0.011 | 0.015 | ORD | MID|PHA02774; | 0.0 |
| 25 | cand_O | P03116 | 409 | LITFINALKLWLKG ++++*++*++ | HHHHHHHHHHHHHC | c2-AT-5 | multi | 0.01 | 0.009 | 0.014 | ORD | MID|PHA02774; | 0.0 |
| 26 | fp_O | P03116 | 412 | FINALKLWLKGIPK +*++*++ | HHHHHHHHHHCCCC | c3-4 | multi-selected | 0.01 | 0.013 | 0.018 | ORD | MID|PHA02774; | 0.0 |
| 27 | fp_O | P03116 | 446 | LIHFLGGSVLSFAN | HHHHHCCCEEECCC | c3-4 | uniq | 0.01 | 0.012 | 0.024 | ORD | MID|PHA02774; | 0.286 |
| 28 | fp_beta_O | P03116 | 514 | KAPPLLVTSNIDVQA | CCCCEEEECCCCCCC | c1b-AT-4 | multi | 0.02 | 0.023 | 0.209 | ORD | MID|PHA02774; | 0.571 |
| 29 | fp_beta_O | P03116 | 515 | APPLLVTSNIDVQA | CCCEEEECCCCCCC | c2-AT-4 | multi-selected | 0.018 | 0.022 | 0.205 | ORD | MID|PHA02774; | 0.571 |
| 30 | fp_beta_O | P03116 | 531 | RYLYLHSRVQTFRFEQ | CCCCCCCEEEEEECCC | c1a-4 | uniq | 0.012 | 0.018 | 0.191 | ORD | MID|PHA02774; | 0.571 |
| 31 | fp_D | P03116 | 567 | FFVRLWGRLDLIDE | HHHHHHHCCCCCCC | c3-4 | uniq | 0.184 | 0.165 | 0.153 | boundary | MID|PHA02774; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment