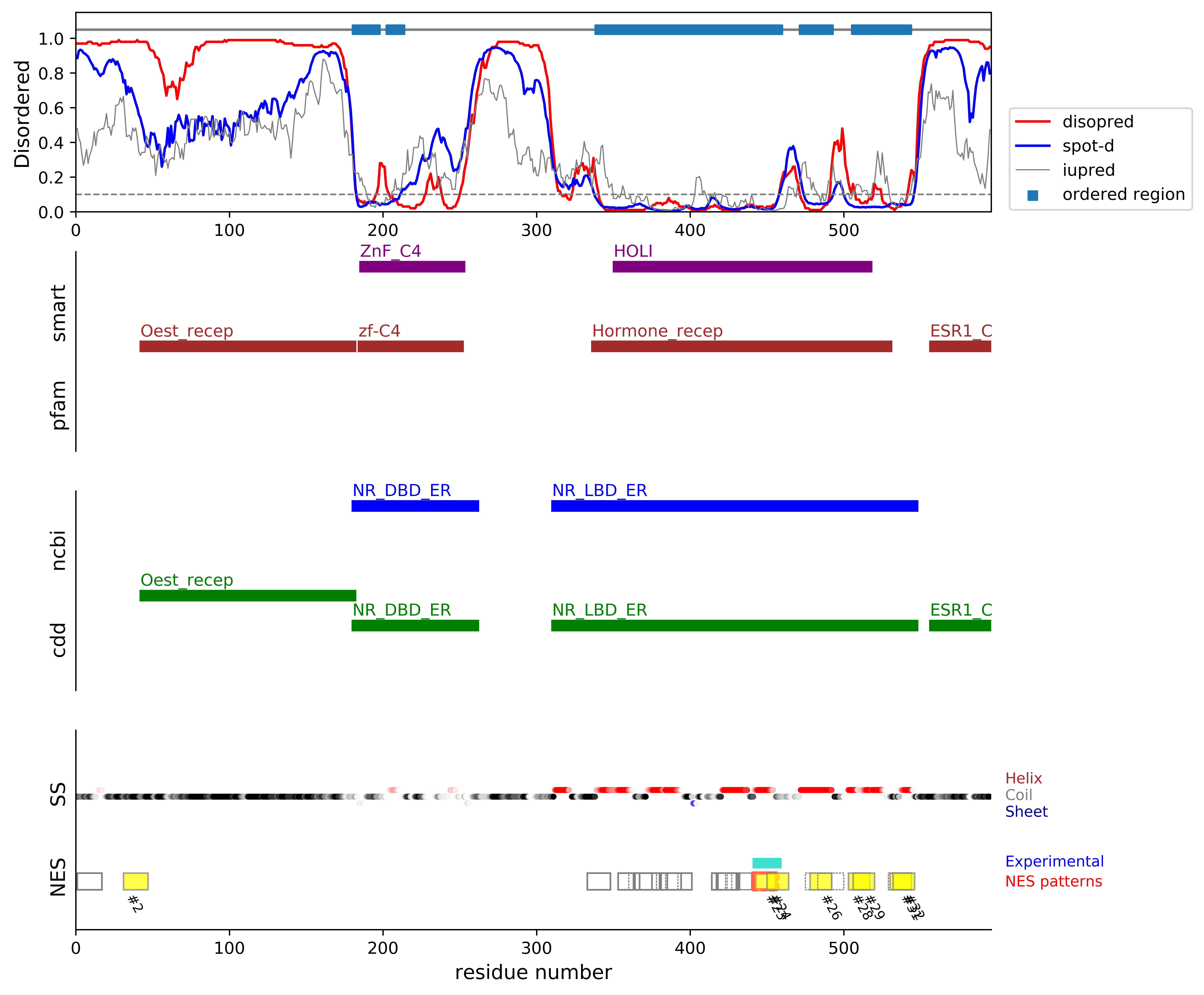

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | P03372 | 1 | MTMTLHTKASGMALLH | CCCCCCCCCCCCCHHH | c1a-AT-4 | uniq | 0.971 | 0.87 | 0.384 | DISO | 0.0 | |
| 2 | fp_D | P03372 | 31 | LKIPLERPLGEVYLDS | CCCCCCCCCCCCCCCC | c1a-4 | uniq | 0.981 | 0.61 | 0.434 | DISO | boundary|Oest_recep; | 0.0 |
| 3 | fp_D | P03372 | 333 | PTRPFSEASMMGLLT | CCCCCCHHHHHHHHH | c1b-AT-4 | uniq | 0.13 | 0.085 | 0.263 | boundary | MID|NR_LBD_ER; | 0.0 |
| 4 | fp_D | P03372 | 353 | ELVHMINWAKRVPG | HHHHHHHHHHHCCC | c3-AT | uniq | 0.01 | 0.029 | 0.085 | boundary | MID|NR_LBD_ER; | 0.0 |
| 5 | fp_O | P03372 | 360 | WAKRVPGFVDLTLHD | HHHHCCCCCCCCHHH | c1b-4 | multi | 0.013 | 0.033 | 0.067 | ORD | MID|NR_LBD_ER; | 0.0 |
| 6 | fp_O | P03372 | 363 | RVPGFVDLTLHD | HCCCCCCCCHHH | c2-rev | multi-selected | 0.013 | 0.034 | 0.065 | ORD | MID|NR_LBD_ER; | 0.0 |
| 7 | fp_O | P03372 | 364 | VPGFVDLTLHDQVHLLE | CCCCCCCCHHHHHHHHH | c1c-4 | multi-selected | 0.024 | 0.026 | 0.05 | ORD | MID|NR_LBD_ER; | 0.0 |
| 8 | fp_O | P03372 | 378 | LLECAWLEILMIGL | HHHHHHHHHHHHHH | c2-AT-5 | multi | 0.054 | 0.007 | 0.013 | ORD | MID|NR_LBD_ER; | 0.0 |
| 9 | fp_O | P03372 | 378 | LLECAWLEILMIGLVW | HHHHHHHHHHHHHHHH | c1a-AT-5 | multi | 0.053 | 0.008 | 0.013 | ORD | MID|NR_LBD_ER; | 0.0 |
| 10 | fp_O | P03372 | 380 | ECAWLEILMIGLVW | HHHHHHHHHHHHHH | c2-4 | multi-selected | 0.053 | 0.007 | 0.013 | ORD | MID|NR_LBD_ER; | 0.0 |
| 11 | fp_O | P03372 | 384 | LEILMIGLVWRSMEHPG | HHHHHHHHHHHHCCCCC | c1cR-4 | multi | 0.044 | 0.014 | 0.019 | ORD | MID|NR_LBD_ER; | 0.0 |
| 12 | fp_O | P03372 | 385 | EILMIGLVWRSMEHPG | HHHHHHHHHHHCCCCC | c1aR-4 | multi-selected | 0.043 | 0.015 | 0.02 | ORD | MID|NR_LBD_ER; | 0.0 |
| 13 | fp_O | P03372 | 414 | QGKCVEGMVEIFDMLL | CCCCCCCHHHHHHHHH | c1a-4 | multi-selected | 0.018 | 0.038 | 0.064 | ORD | MID|NR_LBD_ER; | 0.0 |
| 14 | fp_O | P03372 | 414 | QGKCVEGMVEIFDMLL | CCCCCCCHHHHHHHHH | c1d-4 | multi | 0.018 | 0.038 | 0.064 | ORD | MID|NR_LBD_ER; | 0.0 |
| 15 | fp_O | P03372 | 417 | CVEGMVEIFDMLLA | CCCCHHHHHHHHHH | c3-4 | multi-selected | 0.014 | 0.027 | 0.054 | ORD | MID|NR_LBD_ER; | 0.0 |
| 16 | fp_O | P03372 | 417 | CVEGMVEIFDMLLAT | CCCCHHHHHHHHHHH | c1b-5 | multi | 0.014 | 0.026 | 0.052 | ORD | MID|NR_LBD_ER; | 0.0 |
| 17 | fp_O | P03372 | 418 | VEGMVEIFDMLLAT | CCCHHHHHHHHHHH | c2-4 | multi-selected | 0.013 | 0.023 | 0.051 | ORD | MID|NR_LBD_ER; | 0.0 |
| 18 | fp_O | P03372 | 423 | EIFDMLLATSSRFRMMN | HHHHHHHHHHHHHHHCC | c1c-AT-5 | multi | 0.018 | 0.02 | 0.049 | ORD | MID|NR_LBD_ER; | 0.0 |
| 19 | fp_O | P03372 | 424 | IFDMLLATSSRFRMMN | HHHHHHHHHHHHHHCC | c1d-AT-5 | multi | 0.019 | 0.02 | 0.046 | ORD | MID|NR_LBD_ER; | 0.0 |
| 20 | fp_D | P03372 | 427 | MLLATSSRFRMMNLQG | HHHHHHHHHHHCCCCH | c1a-AT-5 | multi | 0.024 | 0.022 | 0.048 | boundary | MID|NR_LBD_ER; | 0.0 |
| 21 | cand_D | P03372 | 441 | QGEEFVCLKSIILLN ++++++++++++ | CHHHHHHHHHHHHHC | c1b-4 | multi | 0.031 | 0.019 | 0.018 | boundary | MID|NR_LBD_ER; | 0.0 |
| 22 | cand_D | P03372 | 441 | QGEEFVCLKSIILLNS +++++++++++++ | CHHHHHHHHHHHHHCC | c1d-4 | multi | 0.032 | 0.021 | 0.017 | boundary | MID|NR_LBD_ER; | 0.0 |
| 23 | cand_D | P03372 | 442 | GEEFVCLKSIILLN ++++++++++++ | HHHHHHHHHHHHHC | c2-4 | multi-selected | 0.03 | 0.019 | 0.015 | boundary | MID|NR_LBD_ER; | 0.0 |
| 24 | fp_D | P03372 | 450 | SIILLNSGVYTFLS +++++++ | HHHHHCCCCCCCCC | c3-4 | uniq | 0.102 | 0.096 | 0.02 | boundary | MID|NR_LBD_ER; | 0.0 |
| 25 | fp_D | P03372 | 475 | IHRVLDKITDTLIHLMA | HHHHHHHHHHHHHHHHH | c1c-AT-4 | multi | 0.026 | 0.051 | 0.139 | boundary | MID|NR_LBD_ER; | 0.0 |
| 26 | fp_D | P03372 | 478 | VLDKITDTLIHLMA | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.027 | 0.049 | 0.12 | boundary | MID|NR_LBD_ER; | 0.0 |
| 27 | fp_D | P03372 | 483 | TDTLIHLMAKAGLTLQQ | HHHHHHHHHHCCCCCCC | c1c-AT-4 | multi | 0.195 | 0.09 | 0.141 | boundary | MID|NR_LBD_ER; | 0.0 |
| 28 | fp_D | P03372 | 503 | RLAQLLLILSHIRH | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.088 | 0.03 | 0.119 | boundary | MID|NR_LBD_ER; | 0.0 |
| 29 | fp_D | P03372 | 506 | QLLLILSHIRHMSN | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.076 | 0.027 | 0.125 | boundary | MID|NR_LBD_ER; | 0.0 |
| 30 | fp_D | P03372 | 529 | KCKNVVPLYDLLLEM | CCCCCCCCHHHHHHH | c1b-4 | multi | 0.05 | 0.043 | 0.069 | boundary | boundary|NR_LBD_ER; | 0.0 |
| 31 | fp_D | P03372 | 530 | CKNVVPLYDLLLEM | CCCCCCCHHHHHHH | c2-4 | multi-selected | 0.051 | 0.043 | 0.064 | boundary | boundary|NR_LBD_ER; | 0.0 |
| 32 | fp_D | P03372 | 532 | NVVPLYDLLLEMLD | CCCCCHHHHHHHHH | c3-4 | multi-selected | 0.081 | 0.045 | 0.079 | boundary | boundary|NR_LBD_ER; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment