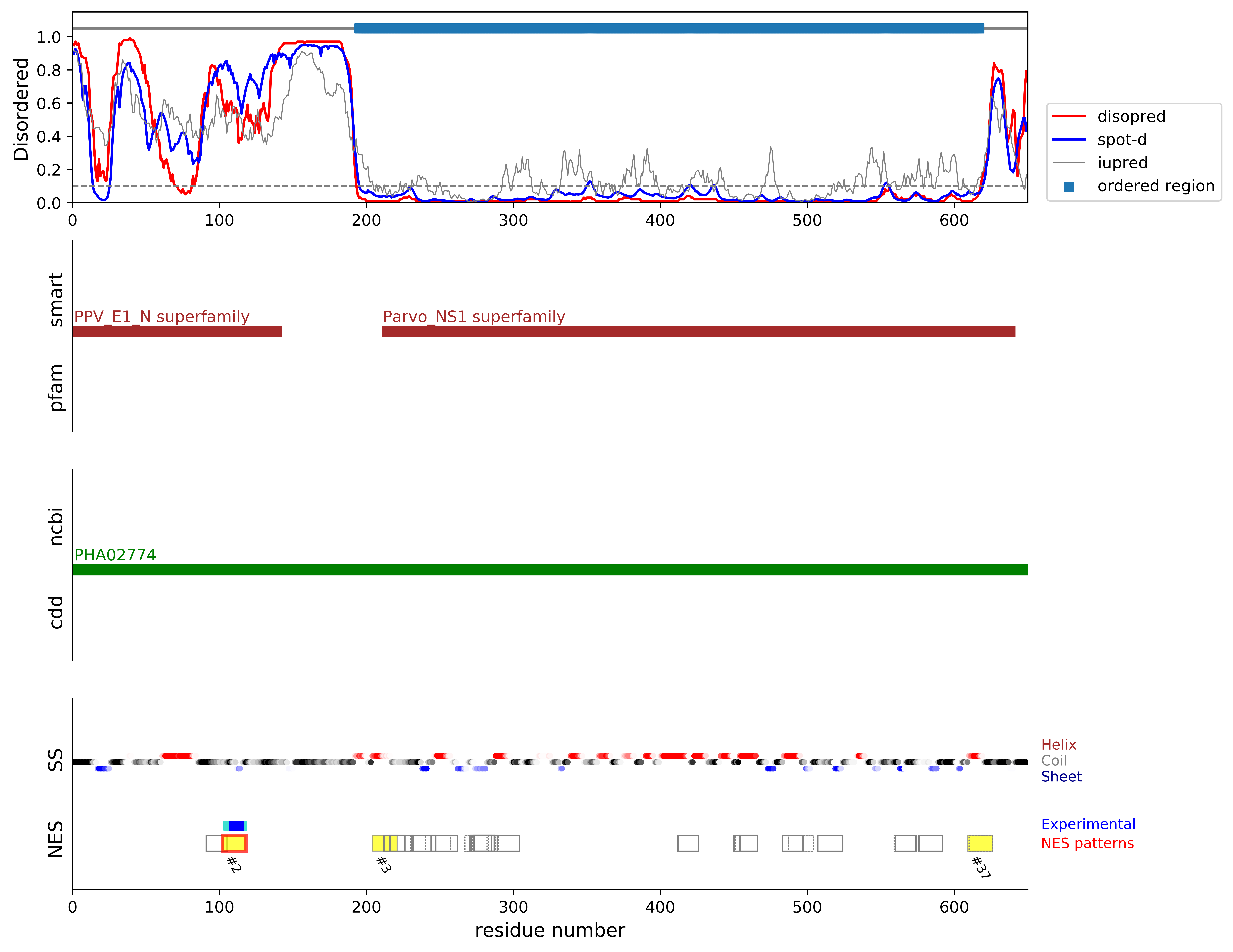

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | P04014 | 91 | YVSPISNVANAVES | CCCCCCCCCCCCCC | c3-AT | uniq | 0.706 | 0.765 | 0.515 | DISO | MID|PHA02774; | 0.0 |
| 2 | cand_D | P04014 | 102 | VESEISPRLDAIKLTT ++++*++*++ | CCCCCCCCCCEEECCC | c1a-4 | uniq | 0.523 | 0.731 | 0.507 | DISO | MID|PHA02774; | 0.143 |
| 3 | fp_D | P04014 | 204 | IRSTLHGKFKDCFGLSF | HHHHHHHHHHHHHCCCC | c1c-4 | uniq | 0.01 | 0.04 | 0.075 | boundary | MID|PHA02774; | 0.0 |
| 4 | fp_O | P04014 | 212 | FKDCFGLSFVDLIR | HHHHHCCCCCCCCC | c2-4 | multi-selected | 0.013 | 0.041 | 0.069 | ORD | MID|PHA02774; | 0.0 |
| 5 | fp_O | P04014 | 212 | FKDCFGLSFVDLIRPFKS | HHHHHCCCCCCCCCCCCC | c4-4 | multi | 0.017 | 0.047 | 0.075 | ORD | MID|PHA02774; | 0.0 |
| 6 | fp_O | P04014 | 216 | FGLSFVDLIRPFKSDR | HCCCCCCCCCCCCCCC | c1aR-4 | multi-selected | 0.02 | 0.055 | 0.08 | ORD | MID|PHA02774; | 0.0 |
| 7 | fp_beta_O | P04014 | 231 | RTTCADWVVAGFGIHH | CCCCCCCEEEECCCCH | c1a-AT-4 | multi-selected | 0.011 | 0.025 | 0.113 | ORD | MID|PHA02774; | 0.571 |
| 8 | fp_beta_O | P04014 | 231 | RTTCADWVVAGFGIHH | CCCCCCCEEEECCCCH | c1d-AT-4 | multi | 0.011 | 0.025 | 0.113 | ORD | MID|PHA02774; | 0.571 |
| 9 | fp_O | P04014 | 240 | AGFGIHHSIADAFQKLI | EECCCCHHHHHHHHHHH | c1cR-5 | multi | 0.009 | 0.014 | 0.081 | ORD | MID|PHA02774; | 0.0 |
| 10 | fp_O | P04014 | 244 | IHHSIADAFQKLIEPLSL | CCHHHHHHHHHHHHCCCC | c4-4 | multi-selected | 0.01 | 0.012 | 0.064 | ORD | MID|PHA02774; | 0.0 |
| 11 | fp_beta_O | P04014 | 267 | WLTNAWGMVLLVLIRF | EECCCCCEEEEEEEEC | c1a-AT-5 | multi | 0.01 | 0.009 | 0.013 | ORD | MID|PHA02774; | 0.571 |
| 12 | fp_beta_O | P04014 | 267 | WLTNAWGMVLLVLIRF | EECCCCCEEEEEEEEC | c1d-AT-5 | multi | 0.01 | 0.009 | 0.013 | ORD | MID|PHA02774; | 0.571 |
| 13 | fp_beta_O | P04014 | 267 | WLTNAWGMVLLVLIR | EECCCCCEEEEEEEE | c1b-AT-5 | multi | 0.01 | 0.009 | 0.013 | ORD | MID|PHA02774; | 0.571 |
| 14 | fp_beta_O | P04014 | 270 | NAWGMVLLVLIRFKVNK | CCCCEEEEEEEECCCCC | c1c-4 | multi-selected | 0.01 | 0.014 | 0.015 | ORD | MID|PHA02774; | 1.0 |
| 15 | fp_beta_O | P04014 | 270 | NAWGMVLLVLIRFKV | CCCCEEEEEEEECCC | c1b-4 | multi | 0.01 | 0.011 | 0.014 | ORD | MID|PHA02774; | 1.0 |
| 16 | fp_beta_O | P04014 | 271 | AWGMVLLVLIRFKVNK | CCCEEEEEEEECCCCC | c1a-4 | multi-selected | 0.01 | 0.014 | 0.015 | ORD | MID|PHA02774; | 1.0 |
| 17 | fp_beta_O | P04014 | 271 | AWGMVLLVLIRFKVNK | CCCEEEEEEEECCCCC | c1d-4 | multi | 0.01 | 0.014 | 0.015 | ORD | MID|PHA02774; | 1.0 |
| 18 | fp_beta_O | P04014 | 271 | AWGMVLLVLIRFKV | CCCEEEEEEEECCC | c2-4 | multi-selected | 0.01 | 0.011 | 0.015 | ORD | MID|PHA02774; | 1.0 |
| 19 | fp_beta_O | P04014 | 272 | WGMVLLVLIRFKVNKSR | CCEEEEEEEECCCCCCH | c1cR-4 | multi | 0.01 | 0.016 | 0.018 | ORD | MID|PHA02774; | 0.75 |
| 20 | fp_beta_O | P04014 | 272 | WGMVLLVLIRFKVNK | CCEEEEEEEECCCCC | c1b-4 | multi | 0.01 | 0.014 | 0.016 | ORD | MID|PHA02774; | 0.857 |
| 21 | fp_beta_O | P04014 | 273 | GMVLLVLIRFKVNK | CEEEEEEEECCCCC | c2-5 | multi-selected | 0.01 | 0.013 | 0.016 | ORD | MID|PHA02774; | 0.857 |
| 22 | fp_beta_O | P04014 | 273 | GMVLLVLIRFKVNKSR | CEEEEEEEECCCCCCH | c1aR-4 | multi-selected | 0.01 | 0.016 | 0.019 | ORD | MID|PHA02774; | 0.714 |
| 23 | fp_O | P04014 | 287 | SRCTVARTLGTLLNIPE | CHHHHHHHHHHHCCCCC | c1c-4 | multi-selected | 0.01 | 0.02 | 0.122 | ORD | MID|PHA02774; | 0.0 |
| 24 | fp_O | P04014 | 288 | RCTVARTLGTLLNIPE | HHHHHHHHHHHCCCCC | c1d-AT-4 | multi | 0.01 | 0.019 | 0.127 | ORD | MID|PHA02774; | 0.0 |
| 25 | fp_O | P04014 | 290 | TVARTLGTLLNIPE | HHHHHHHHHCCCCC | c3-AT | multi | 0.01 | 0.017 | 0.14 | ORD | MID|PHA02774; | 0.0 |
| 26 | fp_O | P04014 | 412 | HYKHAEMKKMSIKQ | HHHHHHHHCCCHHH | c2-AT-4 | uniq | 0.026 | 0.072 | 0.109 | ORD | MID|PHA02774; | 0.0 |
| 27 | fp_O | P04014 | 450 | QNIEFIPFLSKLKLWL | CCCCHHHHHHHHHHHH | c1a-4 | multi-selected | 0.01 | 0.012 | 0.033 | ORD | MID|PHA02774; | 0.0 |
| 28 | fp_O | P04014 | 450 | QNIEFIPFLSKLKLWL | CCCCHHHHHHHHHHHH | c1d-4 | multi | 0.01 | 0.012 | 0.033 | ORD | MID|PHA02774; | 0.0 |
| 29 | fp_O | P04014 | 451 | NIEFIPFLSKLKLWL | CCCHHHHHHHHHHHH | c1b-5 | multi | 0.01 | 0.012 | 0.033 | ORD | MID|PHA02774; | 0.0 |
| 30 | fp_O | P04014 | 454 | FIPFLSKLKLWL | HHHHHHHHHHHH | c2-rev | multi-selected | 0.01 | 0.01 | 0.035 | ORD | MID|PHA02774; | 0.0 |
| 31 | fp_O | P04014 | 483 | GKSCFCMSLIKFLG | CHHHHHHHHHHHHC | c2-4 | multi-selected | 0.01 | 0.012 | 0.028 | ORD | MID|PHA02774; | 0.0 |
| 32 | fp_O | P04014 | 487 | FCMSLIKFLGGTVISYV | HHHHHHHHHCCCEEEEC | c1cR-4 | multi | 0.01 | 0.01 | 0.023 | ORD | MID|PHA02774; | 0.0 |
| 33 | fp_O | P04014 | 507 | SHFWLQPLTDAKVALLD | CCCCCCCCCCCCEEEEE | c1c-AT-4 | uniq | 0.01 | 0.013 | 0.063 | ORD | MID|PHA02774; | 0.0 |
| 34 | fp_beta_O | P04014 | 559 | KCPPLLVTSNIDISK | CCCCEEEECCCCCCC | c1b-AT-4 | multi | 0.031 | 0.023 | 0.124 | ORD | MID|PHA02774; | 0.571 |
| 35 | fp_beta_O | P04014 | 560 | CPPLLVTSNIDISK | CCCEEEECCCCCCC | c2-AT-4 | multi-selected | 0.029 | 0.023 | 0.124 | ORD | MID|PHA02774; | 0.571 |
| 36 | fp_O | P04014 | 576 | KYKYLHSRVTTFTFPN | CCCCCCCCEEEEECCC | c1a-4 | uniq | 0.022 | 0.019 | 0.168 | ORD | MID|PHA02774; | 0.429 |
| 37 | fp_D | P04014 | 609 | NWKCFFERLSSSLDIED | CHHHHHHHHHHCCCCCC | c1c-4 | multi-selected | 0.15 | 0.143 | 0.205 | boundary | MID|PHA02774; | 0.0 |
| 38 | fp_D | P04014 | 610 | WKCFFERLSSSLDIED | HHHHHHHHHHCCCCCC | c1d-4 | multi | 0.159 | 0.148 | 0.213 | boundary | MID|PHA02774; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment