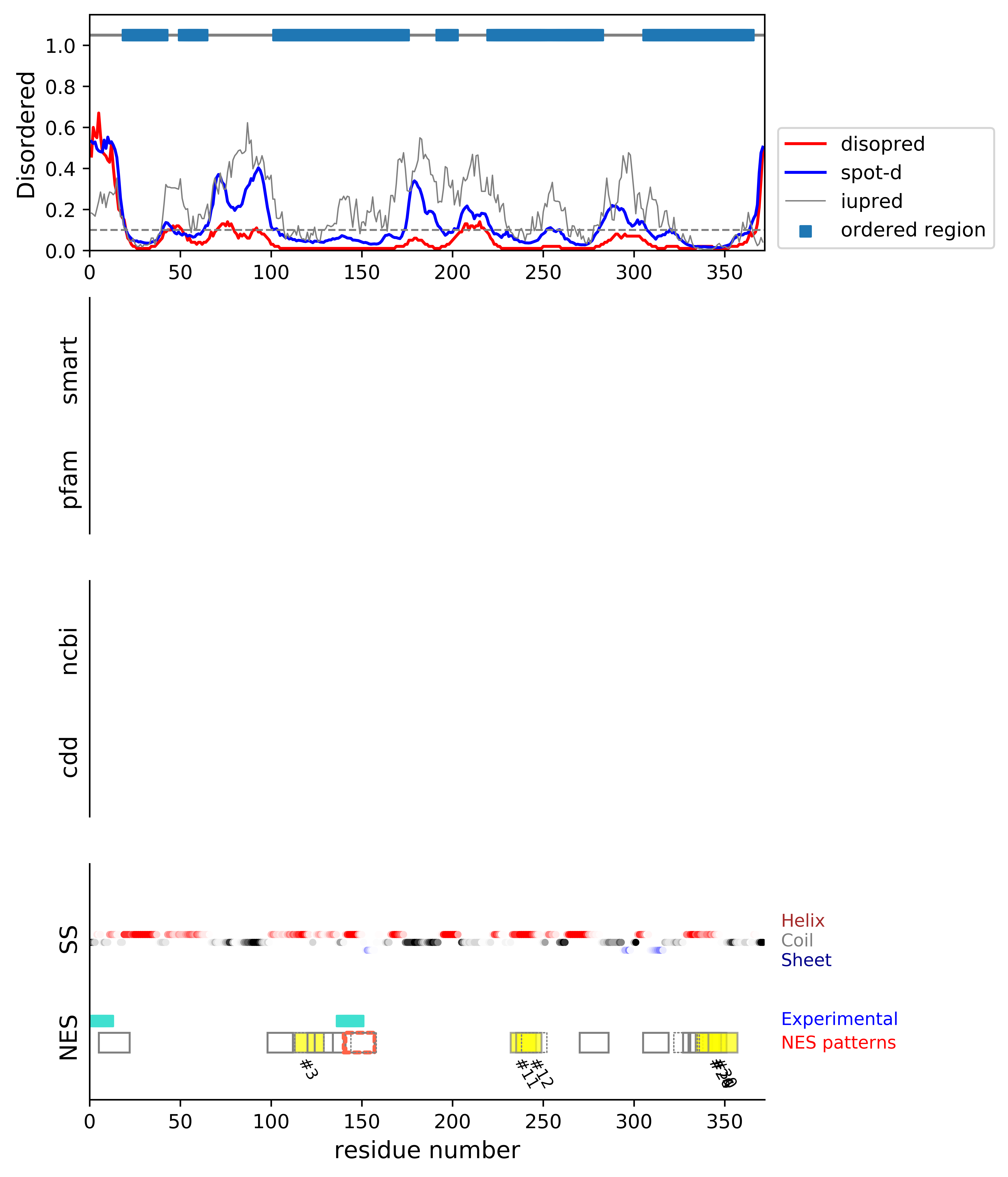

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | P0CA03 | 5 | EELGLSKAARSQVDLNQ ++++++ | HHCCCCHHHHHHCCHHH | c1c-AT-4 | uniq | 0.347 | 0.392 | 0.224 | boundary | 0.0 | |
| 2 | fp_D | P0CA03 | 98 | VDWKAAVSAIELEY | CCHHHHHHHHHHHH | c2-AT-4 | uniq | 0.021 | 0.097 | 0.187 | boundary | 0.0 | |
| 3 | fp_D | P0CA03 | 113 | VKRRFYRALEGLDLYL | HHHHHHHHHCCHHHHH | c1a-4 | multi-selected | 0.01 | 0.045 | 0.075 | boundary | 0.0 | |
| 4 | fp_D | P0CA03 | 113 | VKRRFYRALEGLDLYL | HHHHHHHHHCCHHHHH | c1d-AT-4 | multi | 0.01 | 0.045 | 0.075 | boundary | 0.0 | |
| 5 | fp_O | P0CA03 | 120 | ALEGLDLYLKNITK | HHCCHHHHHHHHHH | c3-4 | multi-selected | 0.01 | 0.045 | 0.087 | ORD | 0.0 | |
| 6 | fp_O | P0CA03 | 124 | LDLYLKNITKTFVNNI + | HHHHHHHHHHHHHCCC | c1aR-4 | multi-selected | 0.01 | 0.051 | 0.126 | ORD | 0.0 | |
| 7 | cand_O | P0CA03 | 140 | DSIQTVQQMLDGVRIIG +++++++++ | CHHHHHHHHHHCEEEEE | c1c-AT-4 | multi | 0.01 | 0.048 | 0.191 | ORD | 0.0 | |
| 8 | fp_O | P0CA03 | 141 | SIQTVQQMLDGVRIIG ++++++++ | HHHHHHHHHHCEEEEE | c1a-5 | multi-selected | 0.01 | 0.046 | 0.186 | ORD | 0.0 | |
| 9 | cand_O | P0CA03 | 141 | SIQTVQQMLDGVRIIG ++++++++ | HHHHHHHHHHCEEEEE | c1d-5 | multi | 0.01 | 0.046 | 0.186 | ORD | 0.0 | |
| 10 | fp_O | P0CA03 | 144 | TVQQMLDGVRIIGR +++++ | HHHHHHHCEEEEEE | c3-4 | multi | 0.01 | 0.042 | 0.17 | ORD | 0.286 | |
| 11 | fp_D | P0CA03 | 232 | NFQALKNIINAFAR | CHHHHHHHHHHHHH | c3-4 | multi-selected | 0.01 | 0.047 | 0.086 | boundary | 0.0 | |
| 12 | fp_D | P0CA03 | 235 | ALKNIINAFARIGD | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.01 | 0.044 | 0.102 | boundary | 0.0 | |
| 13 | fp_D | P0CA03 | 238 | NIINAFARIGDMLG | HHHHHHHHHHHCCC | c3-AT | multi | 0.011 | 0.051 | 0.123 | boundary | 0.0 | |
| 14 | fp_D | P0CA03 | 270 | LLEYIQHSALSVGLKN | HHHHHHHHHHHHCCCC | c1a-AT-5 | uniq | 0.018 | 0.077 | 0.12 | boundary | 0.0 | |
| 15 | fp_beta_D | P0CA03 | 305 | AAQRVYLSTVRVND | HHHHEEEEEEEECC | c2-4 | uniq | 0.024 | 0.085 | 0.202 | boundary | 0.857 | |
| 16 | fp_D | P0CA03 | 322 | TRWETEDVFFTFMLKS | CCCCCCHHHHHHHHHH | c1a-AT-4 | multi | 0.014 | 0.039 | 0.049 | __ | 0.0 | |
| 17 | fp_D | P0CA03 | 322 | TRWETEDVFFTFMLKS | CCCCCCHHHHHHHHHH | c1d-AT-4 | multi | 0.014 | 0.039 | 0.049 | __ | 0.0 | |
| 18 | fp_O | P0CA03 | 327 | EDVFFTFMLKSMAAKI | CHHHHHHHHHHHHHHH | c1aR-4 | multi-selected | 0.015 | 0.023 | 0.026 | ORD | 0.0 | |
| 19 | fp_O | P0CA03 | 330 | FFTFMLKSMAAKIFIVL | HHHHHHHHHHHHHHHHH | c1c-5 | multi-selected | 0.015 | 0.018 | 0.018 | ORD | 0.0 | |
| 20 | fp_D | P0CA03 | 331 | FTFMLKSMAAKIFIVLG | HHHHHHHHHHHHHHHHH | c1c-AT-4 | multi | 0.015 | 0.018 | 0.017 | boundary | 0.0 | |
| 21 | fp_O | P0CA03 | 331 | FTFMLKSMAAKIFIVL | HHHHHHHHHHHHHHHH | c1aR-5 | multi-selected | 0.016 | 0.018 | 0.016 | ORD | 0.0 | |
| 22 | fp_O | P0CA03 | 331 | FTFMLKSMAAKIFIVL | HHHHHHHHHHHHHHHH | c1a-AT-4 | multi | 0.016 | 0.018 | 0.016 | ORD | 0.0 | |
| 23 | fp_O | P0CA03 | 331 | FTFMLKSMAAKIFIVL | HHHHHHHHHHHHHHHH | c1d-4 | multi | 0.016 | 0.018 | 0.016 | ORD | 0.0 | |
| 24 | fp_D | P0CA03 | 334 | MLKSMAAKIFIVLGIYD | HHHHHHHHHHHHHHCCC | c1c-5 | multi-selected | 0.015 | 0.017 | 0.013 | boundary | 0.0 | |
| 25 | fp_D | P0CA03 | 334 | MLKSMAAKIFIVLG | HHHHHHHHHHHHHH | c2-AT-5 | multi | 0.016 | 0.017 | 0.012 | boundary | 0.0 | |
| 26 | fp_D | P0CA03 | 334 | MLKSMAAKIFIVLG | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.016 | 0.017 | 0.012 | boundary | 0.0 | |
| 27 | fp_D | P0CA03 | 335 | LKSMAAKIFIVLGIYD | HHHHHHHHHHHHHCCC | c1a-AT-4 | multi | 0.016 | 0.017 | 0.013 | boundary | 0.0 | |
| 28 | fp_D | P0CA03 | 335 | LKSMAAKIFIVLGIYD | HHHHHHHHHHHHHCCC | c1d-AT-4 | multi | 0.016 | 0.017 | 0.013 | boundary | 0.0 | |
| 29 | fp_D | P0CA03 | 336 | KSMAAKIFIVLGIYD | HHHHHHHHHHHHCCC | c1b-AT-4 | multi | 0.015 | 0.017 | 0.013 | boundary | 0.0 | |
| 30 | fp_D | P0CA03 | 341 | KIFIVLGIYDMFERPE | HHHHHHHCCCCCCCCC | c1aR-4 | multi-selected | 0.014 | 0.025 | 0.024 | boundary | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment