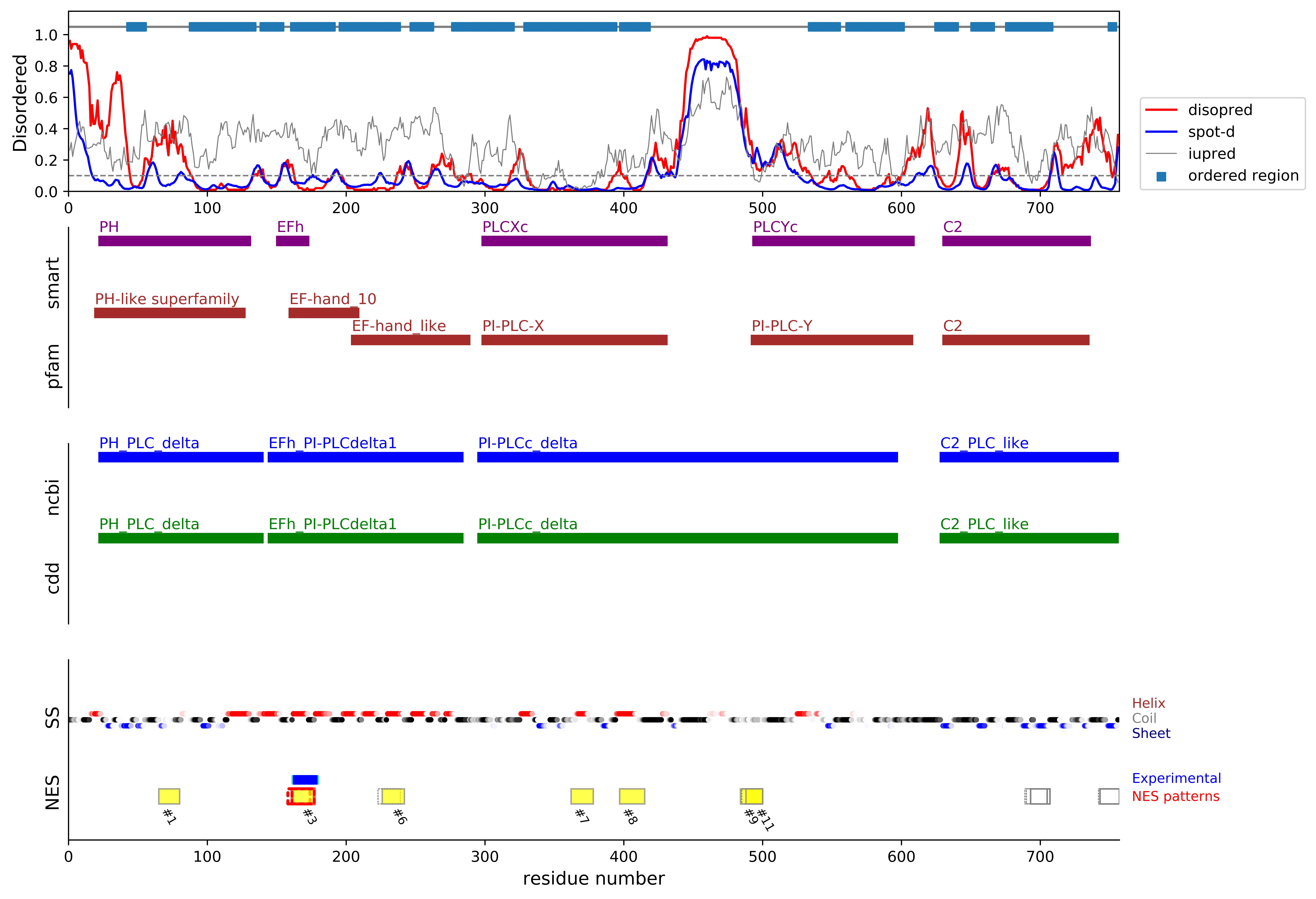

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | P10688 | 65 | QLFSIEDIQEVRMGH | CEEEECCCCCCCCCC | c1b-5 | uniq | 0.328 | 0.054 | 0.374 | boundary | MID|PH_PLC_delta; | 0.143 |
| 2 | cand_D | P10688 | 158 | NKMNFKELKDFLKELN +*+++*++*+ | CCCCHHHHHHHHHHHC | c1aR-5 | multi | 0.069 | 0.067 | 0.307 | boundary | boundary|PH_PLC_delta; boundary|EFh_PI-PLCdelta1; | 0.0 |
| 3 | cand_D | P10688 | 161 | NFKELKDFLKELNIQV +*+++*++*+*+* | CHHHHHHHHHHHCCCC | c1a-5 | multi-selected | 0.039 | 0.064 | 0.268 | boundary | boundary|PH_PLC_delta; boundary|EFh_PI-PLCdelta1; | 0.0 |
| 4 | cand_D | P10688 | 161 | NFKELKDFLKELNIQV +*+++*++*+*+* | CHHHHHHHHHHHCCCC | c1d-5 | multi | 0.039 | 0.064 | 0.268 | boundary | boundary|PH_PLC_delta; boundary|EFh_PI-PLCdelta1; | 0.0 |
| 5 | fp_D | P10688 | 223 | GSAETLSVERLVTFLQ | CCCCCCCHHHHHHHHH | c1d-AT-4 | multi | 0.036 | 0.068 | 0.341 | boundary | MID|EFh_PI-PLCdelta1; | 0.0 |
| 6 | fp_D | P10688 | 226 | ETLSVERLVTFLQHQQ | CCCCHHHHHHHHHHHH | c1aR-4 | multi-selected | 0.057 | 0.061 | 0.358 | boundary | MID|EFh_PI-PLCdelta1; | 0.0 |
| 7 | fp_D | P10688 | 362 | SKILFCDVLRAIRDYA | CCCCHHHHHHHHHCCC | c1aR-4 | uniq | 0.01 | 0.014 | 0.024 | boundary | MID|PI-PLCc_delta; | 0.0 |
| 8 | fp_D | P10688 | 397 | QQRVMARHLRAILGPILL | HHHHHHHHHHHHHHCCCC | c4-4 | uniq | 0.053 | 0.023 | 0.194 | boundary | MID|PI-PLCc_delta; | 0.0 |
| 9 | fp_D | P10688 | 484 | DKLKLVPELSDMIIYC | CCCCCCCCCCCCCCEE | c1a-4 | multi-selected | 0.302 | 0.275 | 0.229 | DISO | MID|PI-PLCc_delta; | 0.0 |
| 10 | fp_D | P10688 | 485 | KLKLVPELSDMIIYC | CCCCCCCCCCCCCEE | c1b-5 | multi | 0.287 | 0.259 | 0.207 | DISO | MID|PI-PLCc_delta; | 0.0 |
| 11 | fp_D | P10688 | 488 | LVPELSDMIIYC | CCCCCCCCCCEE | c2-rev | multi-selected | 0.257 | 0.228 | 0.147 | DISO | MID|PI-PLCc_delta; | 0.0 |
| 12 | fp_beta_D | P10688 | 689 | EFEVTVPDLALVRFMV | EEEEECCEEEEEEEEE | c1a-AT-5 | multi | 0.014 | 0.008 | 0.138 | boundary | MID|C2_PLC_like; | 0.714 |
| 13 | fp_beta_D | P10688 | 690 | FEVTVPDLALVRFMVED | EEEECCEEEEEEEEEEE | c1c-AT-4 | multi | 0.021 | 0.009 | 0.142 | boundary | MID|C2_PLC_like; | 0.75 |
| 14 | fp_beta_D | P10688 | 690 | FEVTVPDLALVRFMV | EEEECCEEEEEEEEE | c1b-4 | multi | 0.015 | 0.008 | 0.136 | boundary | MID|C2_PLC_like; | 0.714 |
| 15 | fp_beta_D | P10688 | 693 | TVPDLALVRFMVED | ECCEEEEEEEEEEE | c2-5 | multi-selected | 0.023 | 0.009 | 0.143 | boundary | MID|C2_PLC_like; | 1.0 |
| 16 | fp_beta_D | P10688 | 693 | TVPDLALVRFMV | ECCEEEEEEEEE | c2-rev | multi-selected | 0.016 | 0.007 | 0.136 | boundary | MID|C2_PLC_like; | 1.0 |
| 17 | fp_beta_D | P10688 | 742 | QHPSATLFVKISIQD | CCCCCCEEEEEEEEC | c1b-AT-4 | multi | 0.255 | 0.054 | 0.257 | DISO | boundary|C2_PLC_like; | 0.714 |
| 18 | fp_beta_D | P10688 | 743 | HPSATLFVKISIQD | CCCCCEEEEEEEEC | c2-AT-4 | multi-selected | 0.245 | 0.054 | 0.256 | DISO | boundary|C2_PLC_like; | 0.714 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment