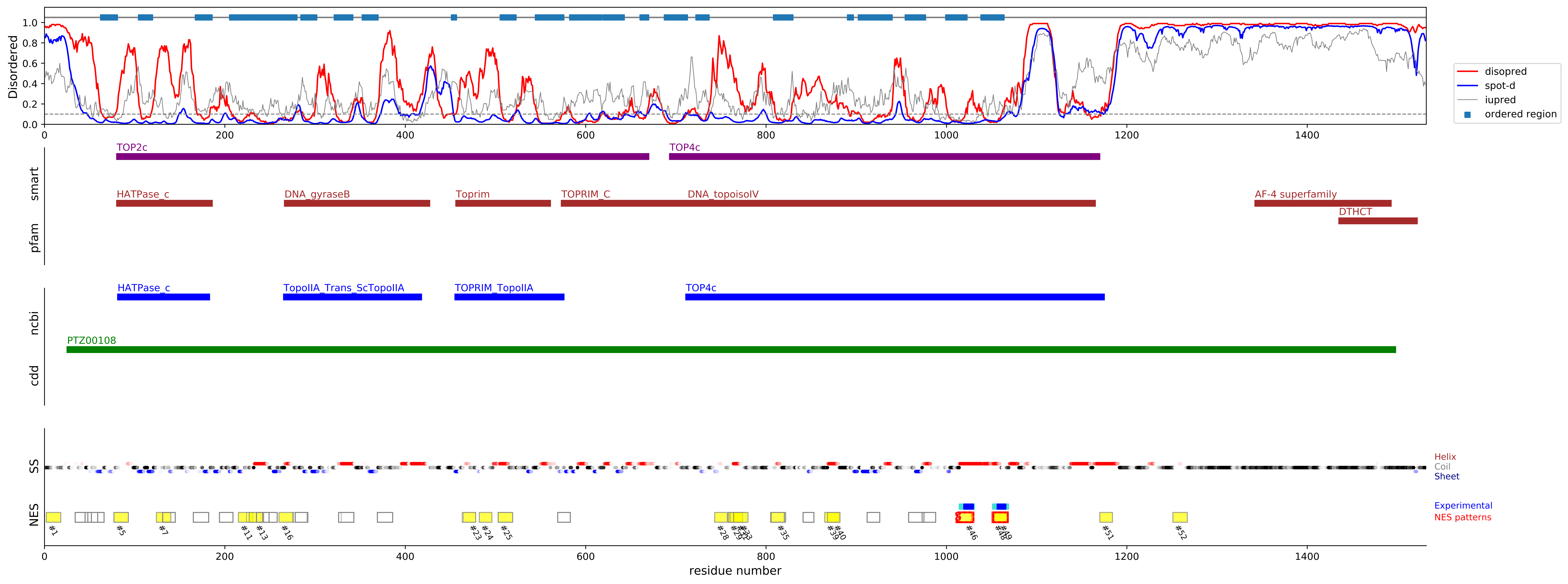

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | P11388 | 2 | EVSPLQPVNENMQVNK | CCCCCCCCCCCCCCCC | c1d-5 | uniq | 0.971 | 0.834 | 0.487 | DISO | boundary|PTZ00108; | 0.0 |
| 2 | fp_D | P11388 | 34 | YQKKTQLEHILLRP | HHHCCCCHHHHCCC | c2-AT-4 | uniq | 0.812 | 0.191 | 0.199 | DISO | boundary|PTZ00108; | 0.0 |
| 3 | fp_D | P11388 | 45 | LRPDTYIGSVELVT | CCCCCCEECCCCEE | c2-AT-4 | uniq | 0.705 | 0.068 | 0.159 | boundary | boundary|PTZ00108; | 0.286 |
| 4 | fp_beta_D | P11388 | 52 | GSVELVTQQMWVYD | ECCCCEEEEEEEEE | c2-AT-4 | uniq | 0.396 | 0.033 | 0.135 | boundary | MID|PTZ00108; | 0.714 |
| 5 | fp_D | P11388 | 77 | FVPGLYKIFDEILVNA | ECCCCCCCCCEEEHHH | c1a-5 | multi-selected | 0.394 | 0.014 | 0.114 | boundary | MID|PTZ00108; | 0.143 |
| 6 | fp_D | P11388 | 77 | FVPGLYKIFDEILVNA | ECCCCCCCCCEEEHHH | c1d-5 | multi | 0.394 | 0.014 | 0.114 | boundary | MID|PTZ00108; | 0.143 |
| 7 | fp_D | P11388 | 124 | GIPVVEHKVEKMYVPA | CCCEEECCCCCCCCCE | c1a-5 | uniq | 0.683 | 0.016 | 0.144 | boundary | MID|PTZ00108; | 0.286 |
| 8 | fp_D | P11388 | 131 | KVEKMYVPALIFGQ | CCCCCCCCEEEEEE | c3-AT | uniq | 0.59 | 0.013 | 0.094 | boundary | MID|PTZ00108; | 0.286 |
| 9 | fp_beta_D | P11388 | 165 | YGAKLCNIFSTKFTVET | CCCEEEEECCCEEEEEE | c1c-4 | uniq | 0.132 | 0.012 | 0.149 | boundary | MID|PTZ00108; | 0.625 |
| 10 | fp_D | P11388 | 194 | WMDNMGRAGEMELKP | ECCCCCCCCCCEEEC | c1b-AT-5 | uniq | 0.21 | 0.064 | 0.334 | boundary | MID|PTZ00108; | 0.0 |
| 11 | fp_D | P11388 | 215 | TCITFQPDLSKFKMQS | EEEEEEECCCCCCCCC | c1a-4 | uniq | 0.065 | 0.026 | 0.158 | boundary | MID|PTZ00108; | 0.429 |
| 12 | fp_D | P11388 | 224 | SKFKMQSLDKDIVALM | CCCCCCCCCHHHHHHH | c1aR-5 | multi | 0.084 | 0.027 | 0.125 | boundary | MID|PTZ00108; | 0.0 |
| 13 | fp_D | P11388 | 227 | KMQSLDKDIVALMVRR | CCCCCCHHHHHHHHHH | c1a-5 | multi-selected | 0.074 | 0.022 | 0.127 | boundary | MID|PTZ00108; | 0.0 |
| 14 | fp_D | P11388 | 235 | IVALMVRRAYDIAG | HHHHHHHHHHHHHC | c3-AT | uniq | 0.036 | 0.009 | 0.116 | __ | MID|PTZ00108; | 0.0 |
| 15 | fp_D | P11388 | 242 | RAYDIAGSTKDVKVFL | HHHHHHCCCCCCEEEE | c1a-AT-4 | uniq | 0.018 | 0.014 | 0.165 | __ | MID|PTZ00108; | 0.0 |
| 16 | fp_D | P11388 | 260 | NKLPVKGFRSYVDMYL | EECCCCCHHHHHHHHH | c1d-4 | uniq | 0.053 | 0.046 | 0.123 | boundary | MID|PTZ00108; | 0.0 |
| 17 | fp_D | P11388 | 275 | LKDKLDETGNSLKVIH | HCCCCCCCCCCCEEEE | c1d-AT-4 | multi | 0.098 | 0.1 | 0.265 | boundary | MID|PTZ00108; | 0.0 |
| 18 | fp_D | P11388 | 278 | KLDETGNSLKVIHE | CCCCCCCCCEEEEE | c3-AT | multi-selected | 0.1 | 0.102 | 0.282 | boundary | MID|PTZ00108; | 0.143 |
| 19 | fp_D | P11388 | 326 | VDYVADQIVTKLVDVVK | HHHHHHHHHHHHHHHHH | c1c-AT-4 | multi-selected | 0.055 | 0.025 | 0.16 | __ | MID|PTZ00108; | 0.0 |
| 20 | fp_D | P11388 | 329 | VADQIVTKLVDVVK | HHHHHHHHHHHHHH | c2-AT-4 | multi | 0.058 | 0.025 | 0.153 | boundary | MID|PTZ00108; | 0.0 |
| 21 | fp_D | P11388 | 369 | ENPTFDSQTKENMTLQP | CCCCCCCCCCCCCCCCC | c1c-AT-4 | uniq | 0.661 | 0.176 | 0.392 | boundary | MID|PTZ00108; | 0.0 |
| 22 | fp_D | P11388 | 463 | DSAKTLAVSGLGVVG | CCHHHHHHCCCCCCC | c1b-AT-4 | multi | 0.419 | 0.063 | 0.176 | DISO | MID|PTZ00108; | 0.0 |
| 23 | fp_D | P11388 | 464 | SAKTLAVSGLGVVG | CHHHHHHCCCCCCC | c2-4 | multi-selected | 0.418 | 0.063 | 0.176 | DISO | MID|PTZ00108; | 0.0 |
| 24 | fp_D | P11388 | 482 | GVFPLRGKILNVRE | EEECCCCCCCCCCC | c3-4 | uniq | 0.602 | 0.038 | 0.176 | boundary | MID|PTZ00108; | 0.0 |
| 25 | fp_D | P11388 | 503 | ENAEINNIIKIVGLQY | HHHHHHHHHHHHCCCC | c1a-4 | multi-selected | 0.119 | 0.043 | 0.185 | boundary | MID|PTZ00108; | 0.0 |
| 26 | fp_D | P11388 | 503 | ENAEINNIIKIVGLQY | HHHHHHHHHHHHCCCC | c1d-4 | multi | 0.119 | 0.043 | 0.185 | boundary | MID|PTZ00108; | 0.0 |
| 27 | fp_beta_D | P11388 | 569 | FLEEFITPIVKVSK | CEEEEECCEEEEEE | c3-4 | uniq | 0.099 | 0.014 | 0.146 | boundary | MID|PTZ00108; | 0.714 |
| 28 | fp_D | P11388 | 743 | KVAQLAGSVAEMSS | EEEEHHHHHHHHCC | c3-4 | uniq | 0.667 | 0.063 | 0.293 | boundary | MID|PTZ00108; | 0.143 |
| 29 | fp_D | P11388 | 758 | HHGEMSLMMTIINLAQ | CCCCCHHHHHHHHHHH | c1a-4 | multi-selected | 0.435 | 0.032 | 0.137 | DISO | MID|PTZ00108; | 0.0 |
| 30 | fp_D | P11388 | 758 | HHGEMSLMMTIINLAQ | CCCCCHHHHHHHHHHH | c1d-4 | multi | 0.435 | 0.032 | 0.137 | DISO | MID|PTZ00108; | 0.0 |
| 31 | fp_D | P11388 | 760 | GEMSLMMTIINLAQ | CCCHHHHHHHHHHH | c2-4 | multi-selected | 0.417 | 0.028 | 0.129 | DISO | MID|PTZ00108; | 0.0 |
| 32 | fp_D | P11388 | 764 | LMMTIINLAQNFVG | HHHHHHHHHHCCCC | c3-AT | multi | 0.296 | 0.023 | 0.11 | DISO | MID|PTZ00108; | 0.0 |
| 33 | fp_D | P11388 | 764 | LMMTIINLAQNFVGSN | HHHHHHHHHHCCCCCC | c1aR-4 | multi-selected | 0.278 | 0.024 | 0.113 | DISO | MID|PTZ00108; | 0.0 |
| 34 | fp_D | P11388 | 805 | YIFTMLSSLARLLFPPK | CCCCCCHHHHHCCCCCC | c1cR-4 | multi | 0.11 | 0.018 | 0.116 | __ | MID|PTZ00108; | 0.0 |
| 35 | fp_D | P11388 | 805 | YIFTMLSSLARLLFPP | CCCCCCHHHHHCCCCC | c1a-5 | multi-selected | 0.114 | 0.018 | 0.116 | boundary | MID|PTZ00108; | 0.0 |
| 36 | fp_D | P11388 | 806 | IFTMLSSLARLLFP | CCCCCHHHHHCCCC | c3-AT | multi | 0.111 | 0.017 | 0.109 | boundary | MID|PTZ00108; | 0.0 |
| 37 | fp_D | P11388 | 806 | IFTMLSSLARLLFPP | CCCCCHHHHHCCCCC | c1b-5 | multi | 0.107 | 0.017 | 0.106 | boundary | MID|PTZ00108; | 0.0 |
| 38 | fp_beta_D | P11388 | 841 | YIPIIPMVLING | CCCCCCEEEECC | c2-rev | uniq | 0.354 | 0.012 | 0.092 | boundary | MID|PTZ00108; | 0.5 |
| 39 | fp_D | P11388 | 865 | PNFDVREIVNNIRRLM | CCCCHHHHHHHHHHHH | c1aR-5 | multi-selected | 0.233 | 0.023 | 0.251 | DISO | MID|PTZ00108; | 0.0 |
| 40 | fp_D | P11388 | 868 | DVREIVNNIRRLMD | CHHHHHHHHHHHHC | c3-4 | multi-selected | 0.215 | 0.022 | 0.276 | DISO | MID|PTZ00108; | 0.0 |
| 41 | fp_beta_D | P11388 | 912 | EVAILNSTTIEISE | EEEEECCCEEEEEE | c3-AT | uniq | 0.043 | 0.017 | 0.181 | boundary | MID|PTZ00108; | 0.571 |
| 42 | fp_beta_D | P11388 | 958 | REYHTDTTVKFVVKMTE | CCCCCCCEEEEEEEECH | c1c-AT-4 | uniq | 0.05 | 0.03 | 0.242 | boundary | MID|PTZ00108; | 0.625 |
| 43 | fp_D | P11388 | 973 | TEEKLAEAERVGLHK | CHHHHHHHHHCCCCC | c1b-AT-4 | uniq | 0.207 | 0.048 | 0.186 | boundary | MID|PTZ00108; | 0.0 |
| 44 | cand_D | P11388 | 1011 | KYDTVLDILRDFFELRL +++++*++*+* | CCCCHHHHHHHHHHHHH | c1c-4 | multi | 0.101 | 0.014 | 0.039 | boundary | MID|PTZ00108; | 0.0 |
| 45 | cand_D | P11388 | 1012 | YDTVLDILRDFFELRL +++++*++*+* | CCCHHHHHHHHHHHHH | c1d-4 | multi | 0.105 | 0.013 | 0.039 | boundary | MID|PTZ00108; | 0.0 |
| 46 | cand_D | P11388 | 1014 | TVLDILRDFFELRLKY +++++*++*+*+ | CHHHHHHHHHHHHHHH | c1a-5 | multi-selected | 0.14 | 0.012 | 0.037 | boundary | MID|PTZ00108; | 0.0 |
| 47 | cand_D | P11388 | 1015 | VLDILRDFFELRLKY +++++*++*+*+ | HHHHHHHHHHHHHHH | c1b-5 | multi | 0.147 | 0.011 | 0.035 | boundary | MID|PTZ00108; | 0.0 |
| 48 | cand_D | P11388 | 1051 | QARFILEKIDGKIIIEN +++++*+++*+++ | HHHHHHHHHCCCCCCCC | c1c-4 | multi-selected | 0.092 | 0.051 | 0.175 | boundary | MID|PTZ00108; | 0.0 |
| 49 | cand_D | P11388 | 1052 | ARFILEKIDGKIIIEN +++++*+++*+++ | HHHHHHHHCCCCCCCC | c1aR-4 | multi-selected | 0.096 | 0.052 | 0.172 | boundary | MID|PTZ00108; | 0.0 |
| 50 | cand_D | P11388 | 1052 | ARFILEKIDGKIIIEN +++++*+++*+++ | HHHHHHHHCCCCCCCC | c1d-4 | multi | 0.096 | 0.052 | 0.172 | boundary | MID|PTZ00108; | 0.0 |
| 51 | fp_D | P11388 | 1170 | DLATFIEELEAVEA | HHHHHHHHHHHHHH | c3-4 | uniq | 0.204 | 0.145 | 0.482 | DISO | MID|PTZ00108; | 0.0 |
| 52 | fp_D | P11388 | 1251 | DGVELEGLKQRLEKKQ | CCCCCCHHHHHHHCCC | c1aR-4 | uniq | 0.973 | 0.909 | 0.773 | DISO | MID|PTZ00108; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment