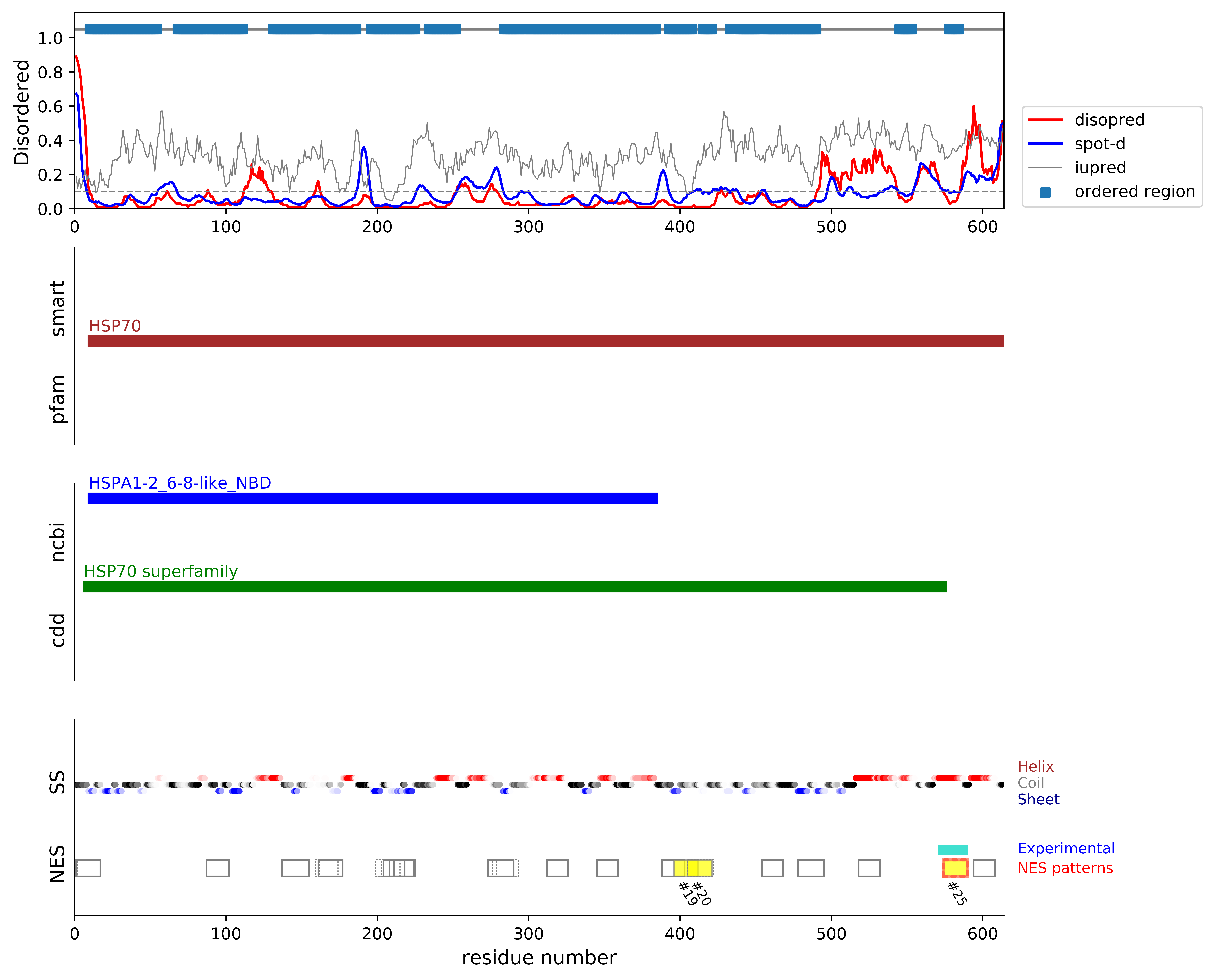

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | P11484 | 1 | MAEGVFQGAIGIDLGT | CCCCCCCCEEEEECCC | c1a-AT-4 | multi-selected | 0.361 | 0.202 | 0.153 | boundary | boundary|HSP70 superfamily; | 0.429 |
| 2 | fp_beta_D | P11484 | 2 | AEGVFQGAIGIDLGT | CCCCCCCEEEEECCC | c1b-AT-4 | multi | 0.325 | 0.171 | 0.151 | boundary | boundary|HSP70 superfamily; | 0.571 |
| 3 | fp_beta_D | P11484 | 87 | KDMKTWPFKVIDVDG | CCCCCCCEEEECCCC | c1b-AT-4 | uniq | 0.047 | 0.04 | 0.271 | boundary | MID|HSP70 superfamily; | 0.571 |
| 4 | fp_beta_D | P11484 | 137 | IGKKVEKAVITVPAYFND | CCCCCCCEEEECCCCCCC | c4-4 | uniq | 0.015 | 0.04 | 0.234 | boundary | MID|HSP70 superfamily; | 0.5 |
| 5 | fp_D | P11484 | 159 | ATKDAGAISGLNVLR | HHCCCCHHCCCEEEE | c1b-AT-4 | multi | 0.052 | 0.05 | 0.277 | boundary | MID|HSP70 superfamily; | 0.0 |
| 6 | fp_D | P11484 | 161 | KDAGAISGLNVLRIIN | CCCCHHCCCEEEEECC | c1a-AT-4 | multi-selected | 0.036 | 0.041 | 0.241 | boundary | MID|HSP70 superfamily; | 0.286 |
| 7 | fp_D | P11484 | 162 | DAGAISGLNVLRIIN | CCCHHCCCEEEEECC | c1b-4 | multi | 0.027 | 0.039 | 0.238 | boundary | MID|HSP70 superfamily; | 0.429 |
| 8 | fp_D | P11484 | 199 | LIFDLGGGTFDVSLLH | EEEECCCCCCEEEEEE | c1a-AT-5 | multi | 0.01 | 0.017 | 0.119 | boundary | MID|HSP70 superfamily; | 0.143 |
| 9 | fp_beta_D | P11484 | 203 | LGGGTFDVSLLHIAG | CCCCCCEEEEEEEEC | c1b-AT-4 | multi | 0.01 | 0.018 | 0.093 | boundary | MID|HSP70 superfamily; | 0.714 |
| 10 | fp_beta_D | P11484 | 204 | GGGTFDVSLLHIAG | CCCCCEEEEEEEEC | c2-4 | multi-selected | 0.01 | 0.018 | 0.091 | boundary | MID|HSP70 superfamily; | 0.714 |
| 11 | fp_beta_D | P11484 | 208 | FDVSLLHIAGGVYTVK | CEEEEEEEECCEEEEE | c1aR-5 | multi-selected | 0.01 | 0.024 | 0.15 | boundary | MID|HSP70 superfamily; | 0.714 |
| 12 | fp_beta_D | P11484 | 211 | SLLHIAGGVYTVKS | EEEEEECCEEEEEC | c3-4 | multi-selected | 0.01 | 0.028 | 0.19 | boundary | MID|HSP70 superfamily; | 0.714 |
| 13 | fp_D | P11484 | 273 | AKRTLSSVTQTTVEVDS | HHHCCCCCCEEEEEEEC | c1c-AT-4 | multi-selected | 0.072 | 0.141 | 0.377 | boundary | MID|HSP70 superfamily; | 0.375 |
| 14 | fp_beta_D | P11484 | 276 | TLSSVTQTTVEVDS | CCCCCCEEEEEEEC | c3-AT | multi | 0.063 | 0.138 | 0.367 | boundary | MID|HSP70 superfamily; | 0.571 |
| 15 | fp_beta_D | P11484 | 279 | SVTQTTVEVDSLFD | CCCEEEEEEECCCC | c3-AT | multi | 0.042 | 0.105 | 0.341 | boundary | MID|HSP70 superfamily; | 1.0 |
| 16 | fp_O | P11484 | 312 | LFKSTLEPVEQVLK | HHHHHHHHHHHHHH | c3-AT | uniq | 0.03 | 0.026 | 0.259 | ORD | MID|HSP70 superfamily; | 0.0 |
| 17 | fp_O | P11484 | 345 | RIPKVQKLLSDFFD | CCHHHHHHHHHHCC | c3-4 | uniq | 0.039 | 0.044 | 0.223 | ORD | MID|HSP70 superfamily; | 0.0 |
| 18 | fp_beta_D | P11484 | 388 | TSDETKDLLLLDVAP | CCCCCCCEEEEECCC | c1b-AT-4 | uniq | 0.023 | 0.096 | 0.262 | boundary | MID|HSP70 superfamily; | 0.571 |
| 19 | fp_D | P11484 | 396 | LLLDVAPLSLGVGMQG | EEEECCCCCCCCCCCC | c1d-5 | uniq | 0.011 | 0.068 | 0.172 | boundary | MID|HSP70 superfamily; | 0.0 |
| 20 | fp_D | P11484 | 405 | LGVGMQGDMFGIVVPR | CCCCCCCCCEEECCCC | c1a-4 | multi-selected | 0.011 | 0.098 | 0.24 | boundary | MID|HSP70 superfamily; | 0.286 |
| 21 | fp_D | P11484 | 405 | LGVGMQGDMFGIVVPRN | CCCCCCCCCEEECCCCC | c1cR-4 | multi | 0.011 | 0.098 | 0.249 | boundary | MID|HSP70 superfamily; | 0.375 |
| 22 | fp_D | P11484 | 454 | CKENTLLGEFDLKN | CCCCCCCCCCCCCC | c2-AT-4 | uniq | 0.038 | 0.061 | 0.354 | __ | MID|HSP70 superfamily; | 0.0 |
| 23 | fp_D | P11484 | 478 | LEAIFEVDANGILKVTA | EEEEEEECCCCCEEEEE | c1c-AT-4 | uniq | 0.064 | 0.032 | 0.26 | boundary | MID|HSP70 superfamily; | 0.375 |
| 24 | fp_D | P11484 | 518 | EIEKMVNQAEEFKA | HHHHHHHHHHHHHH | c3-AT | uniq | 0.263 | 0.074 | 0.435 | boundary | MID|HSP70 superfamily; | 0.0 |
| 25 | cand_D | P11484 | 574 | IEAALSDALAALQIED ++++++++++++++ | HHHHHHHHHHHHHHCC | c1a-4 | multi-selected | 0.104 | 0.115 | 0.347 | boundary | boundary|HSP70 superfamily; | 0.0 |
| 26 | cand_D | P11484 | 574 | IEAALSDALAALQIED ++++++++++++++ | HHHHHHHHHHHHHHCC | c1d-AT-4 | multi | 0.104 | 0.115 | 0.347 | boundary | boundary|HSP70 superfamily; | 0.0 |
| 27 | fp_D | P11484 | 594 | ELRKAEVGLKRVVT | HHHHHHHHHHHHHH | c3-AT | uniq | 0.328 | 0.178 | 0.417 | boundary | boundary|HSP70 superfamily; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment