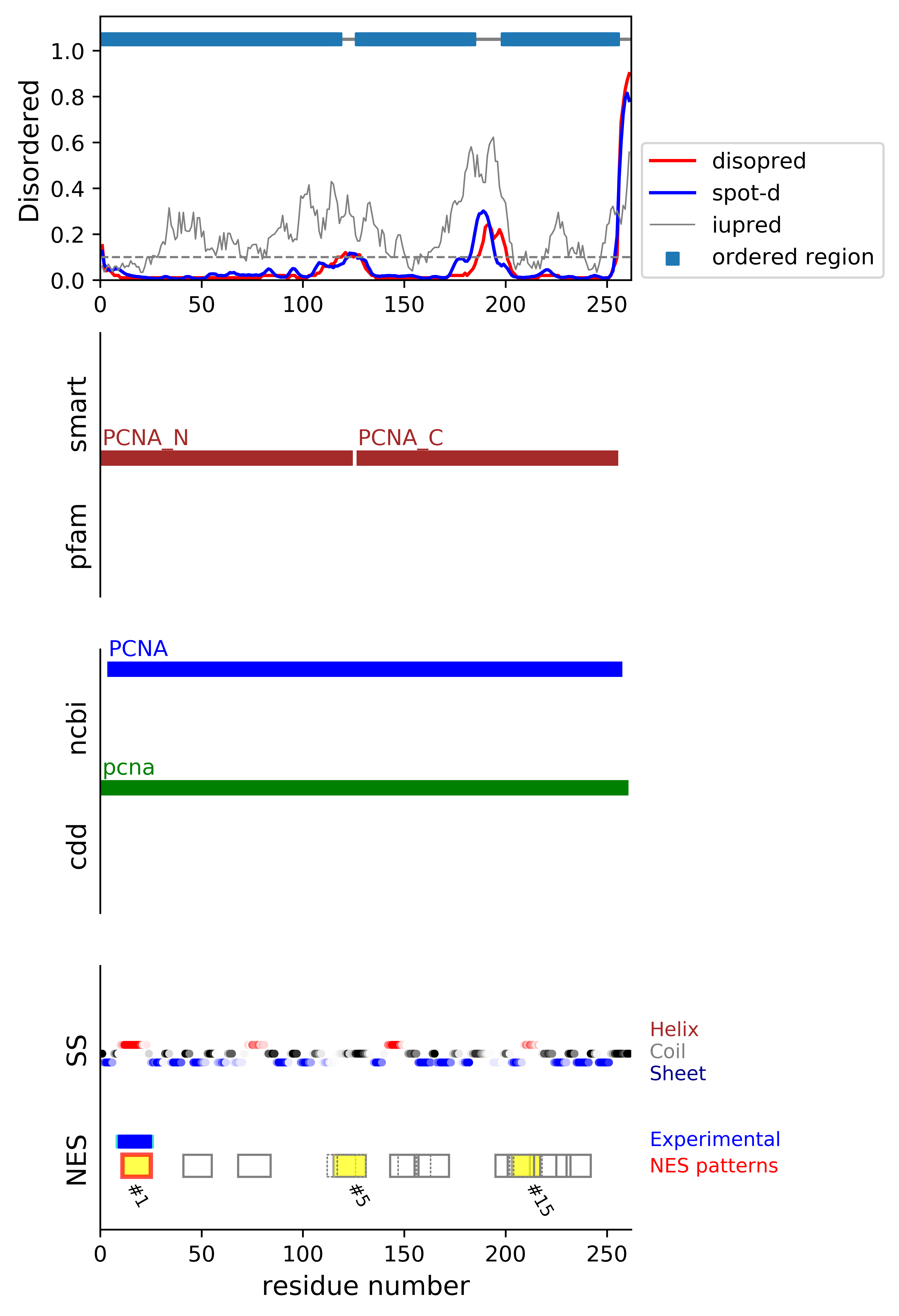

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | cand_D | P12004 | 11 | ILKKVLEALKDLIN +*+++*++*++*+ | HHHHHHHHHHHHHC | c3-4 | uniq | 0.01 | 0.018 | 0.065 | terminal | boundary|pcna; | 0.0 |
| 2 | fp_beta_O | P12004 | 41 | DSSHVSLVQLTLRS | CCCCEEEEEEEECC | c2-4 | uniq | 0.01 | 0.011 | 0.213 | ORD | MID|pcna; | 0.857 |
| 3 | fp_O | P12004 | 68 | MGVNLTSMSKILKCAG | EEECHHHHHHHHHHCC | c1aR-4 | uniq | 0.012 | 0.026 | 0.13 | ORD | MID|pcna; | 0.0 |
| 4 | fp_D | P12004 | 112 | SDYEMKLMDLDVEQ | EEEEECCCCCCCCC | c2-4 | multi | 0.093 | 0.079 | 0.319 | boundary | MID|pcna; | 0.286 |
| 5 | fp_D | P12004 | 115 | EMKLMDLDVEQLGIPE | EECCCCCCCCCCCCCC | c1a-5 | multi-selected | 0.1 | 0.088 | 0.278 | boundary | MID|pcna; | 0.0 |
| 6 | fp_D | P12004 | 117 | KLMDLDVEQLGIPE | CCCCCCCCCCCCCC | c2-5 | multi | 0.104 | 0.092 | 0.261 | boundary | MID|pcna; | 0.0 |
| 7 | fp_O | P12004 | 143 | EFARICRDLSHIGD | HHHHHHHHHCCCCC | c3-4 | multi-selected | 0.01 | 0.018 | 0.112 | ORD | MID|pcna; | 0.0 |
| 8 | fp_O | P12004 | 147 | ICRDLSHIGDAVVISC | HHHHHCCCCCEEEEEE | c1d-4 | multi | 0.01 | 0.014 | 0.105 | ORD | MID|pcna; | 0.143 |
| 9 | fp_beta_D | P12004 | 155 | GDAVVISCAKDGVKFSA | CCEEEEEEECCEEEEEE | c1c-AT-4 | multi-selected | 0.01 | 0.012 | 0.138 | boundary | MID|pcna; | 0.75 |
| 10 | fp_beta_D | P12004 | 156 | DAVVISCAKDGVKFSA | CEEEEEEECCEEEEEE | c1d-AT-4 | multi | 0.01 | 0.011 | 0.141 | boundary | MID|pcna; | 0.714 |
| 11 | fp_beta_D | P12004 | 195 | VTIEMNEPVQLTFALRY | EEEECCCCEEEEEEHHH | c1c-4 | uniq | 0.084 | 0.038 | 0.219 | boundary | MID|pcna; | 0.5 |
| 12 | fp_beta_D | P12004 | 201 | EPVQLTFALRYLNFFT | CCEEEEEEHHHHHHHH | c1a-4 | multi-selected | 0.023 | 0.018 | 0.097 | boundary | MID|pcna; | 0.571 |
| 13 | fp_beta_D | P12004 | 201 | EPVQLTFALRYLNFFT | CCEEEEEEHHHHHHHH | c1d-AT-4 | multi | 0.023 | 0.018 | 0.097 | boundary | MID|pcna; | 0.571 |
| 14 | fp_D | P12004 | 202 | PVQLTFALRYLNFFT | CEEEEEEHHHHHHHH | c1b-AT-5 | multi | 0.018 | 0.016 | 0.086 | boundary | MID|pcna; | 0.429 |
| 15 | fp_D | P12004 | 203 | VQLTFALRYLNFFT | EEEEEEHHHHHHHH | c2-4 | multi-selected | 0.014 | 0.015 | 0.08 | boundary | MID|pcna; | 0.429 |
| 16 | fp_D | P12004 | 204 | QLTFALRYLNFFTK | EEEEEHHHHHHHHH | c3-AT | multi | 0.013 | 0.015 | 0.072 | boundary | MID|pcna; | 0.286 |
| 17 | fp_D | P12004 | 214 | FFTKATPLSSTVTLSM | HHHHCCCCCCEEEEEE | c1d-AT-5 | multi-selected | 0.016 | 0.025 | 0.159 | __ | MID|pcna; | 0.143 |
| 18 | fp_beta_O | P12004 | 217 | KATPLSSTVTLSMSA | HCCCCCCEEEEEEEC | c1b-AT-4 | multi-selected | 0.017 | 0.025 | 0.175 | ORD | MID|pcna; | 0.571 |
| 19 | fp_beta_D | P12004 | 225 | VTLSMSADVPLVVEYKI | EEEEEECCCCEEEEEEE | c1cR-4 | uniq | 0.011 | 0.011 | 0.156 | boundary | boundary|pcna; | 0.5 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment