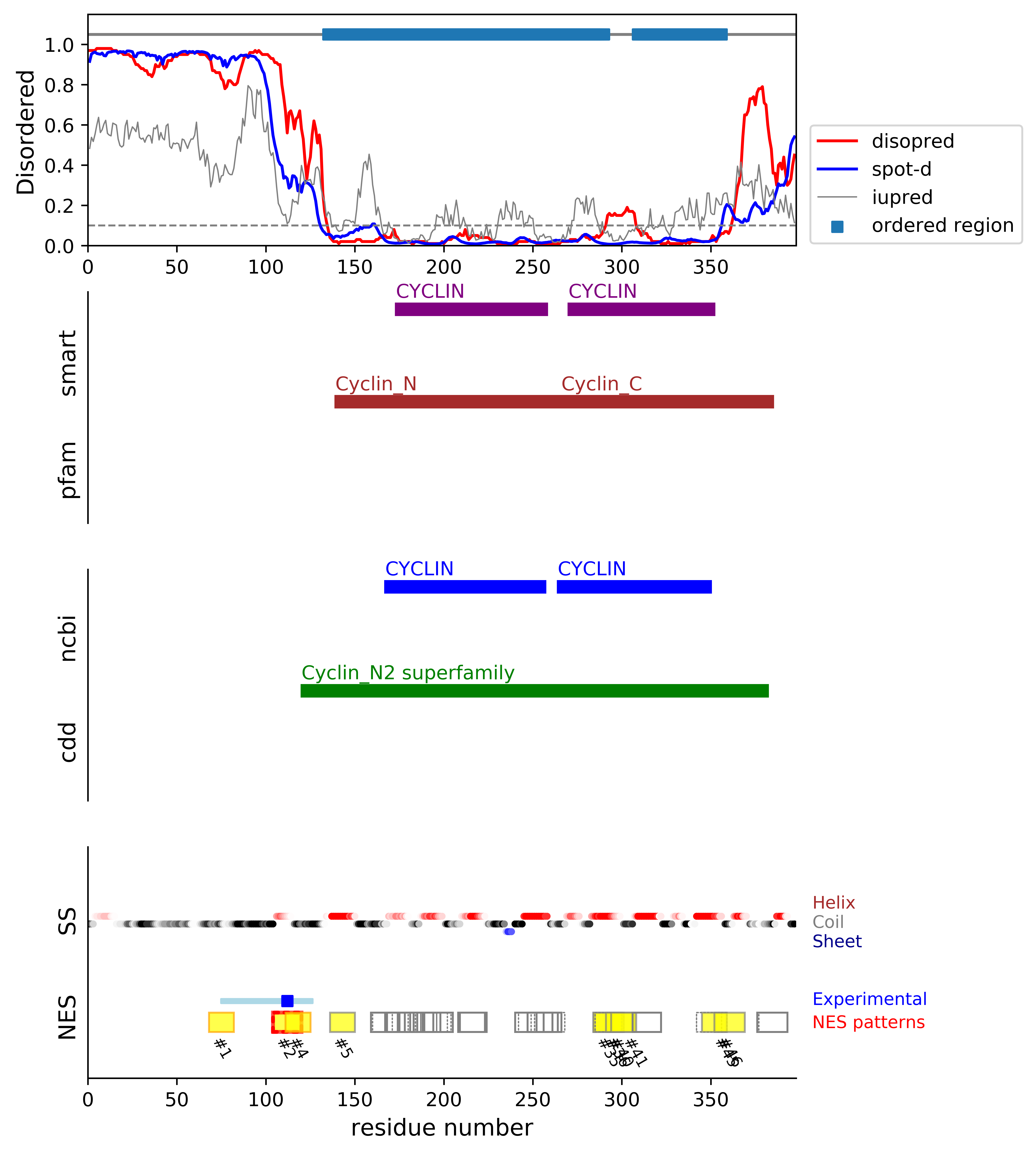

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | P13350 | 68 | PVDKLLEPLKVIEE ...... | CCCCCCCCCCCCCC | c3-4 | uniq | 0.846 | 0.925 | 0.361 | DISO | 0.0 | |
| 2 | cand_D | P13350 | 104 | LPDELCQAFSDVLIHV ........*....... | CHHHHHHHHHHCCCCC | c1a-4 | multi-selected | 0.738 | 0.371 | 0.223 | __ | boundary|Cyclin_N2 superfamily; | 0.0 |
| 3 | cand_D | P13350 | 104 | LPDELCQAFSDVLIHV ........*....... | CHHHHHHHHHHCCCCC | c1d-AT-4 | multi | 0.738 | 0.371 | 0.223 | __ | boundary|Cyclin_N2 superfamily; | 0.0 |
| 4 | fp_D | P13350 | 111 | AFSDVLIHVKDVDA .*............ | HHHHCCCCCCCCCC | c3-4 | uniq | 0.57 | 0.312 | 0.243 | boundary | boundary|Cyclin_N2 superfamily; | 0.0 |
| 5 | fp_D | P13350 | 136 | YVKDIYAYLRSLED | HHHHHHHHHHHHHH | c3-4 | uniq | 0.021 | 0.051 | 0.106 | boundary | boundary|Cyclin_N2 superfamily; | 0.0 |
| 6 | fp_O | P13350 | 159 | HGQEVTGNMRAILIDW | CCCCCCCCCCHHHHHH | c1a-4 | multi-selected | 0.041 | 0.047 | 0.133 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 7 | fp_O | P13350 | 160 | GQEVTGNMRAILIDW | CCCCCCCCCHHHHHH | c1b-AT-4 | multi | 0.042 | 0.043 | 0.115 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 8 | fp_O | P13350 | 167 | MRAILIDWLVQVQMKF | CCHHHHHHHHHHHHCC | c1a-4 | multi-selected | 0.034 | 0.016 | 0.044 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 9 | fp_O | P13350 | 168 | RAILIDWLVQVQMKFRL | CHHHHHHHHHHHHCCCC | c1c-4 | multi-selected | 0.031 | 0.015 | 0.042 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 10 | fp_O | P13350 | 168 | RAILIDWLVQVQMKF | CHHHHHHHHHHHHCC | c1b-4 | multi | 0.033 | 0.015 | 0.043 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 11 | fp_O | P13350 | 171 | LIDWLVQVQMKFRLLQ | HHHHHHHHHHCCCCCC | c1d-5 | multi | 0.028 | 0.015 | 0.031 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 12 | fp_O | P13350 | 174 | WLVQVQMKFRLLQE | HHHHHHHCCCCCCH | c3-4 | multi-selected | 0.02 | 0.016 | 0.028 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 13 | fp_O | P13350 | 178 | VQMKFRLLQETMFMTV | HHHCCCCCCHHHHHHH | c1aR-4 | multi-selected | 0.017 | 0.014 | 0.033 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 14 | fp_O | P13350 | 178 | VQMKFRLLQETMFMTV | HHHCCCCCCHHHHHHH | c1d-4 | multi | 0.017 | 0.014 | 0.033 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 15 | fp_O | P13350 | 180 | MKFRLLQETMFMTVGI | HCCCCCCHHHHHHHHH | c1a-AT-4 | multi | 0.016 | 0.013 | 0.039 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 16 | fp_O | P13350 | 181 | KFRLLQETMFMTVGIID | CCCCCCHHHHHHHHHHH | c1c-5 | multi-selected | 0.015 | 0.013 | 0.054 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 17 | fp_O | P13350 | 181 | KFRLLQETMFMTVGI | CCCCCCHHHHHHHHH | c1b-AT-5 | multi | 0.015 | 0.013 | 0.041 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 18 | fp_O | P13350 | 184 | LLQETMFMTVGIID | CCCHHHHHHHHHHH | c2-AT-5 | multi | 0.015 | 0.011 | 0.059 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 19 | fp_O | P13350 | 187 | ETMFMTVGIIDRFLQEH | HHHHHHHHHHHHHCCCC | c1cR-4 | multi | 0.016 | 0.014 | 0.094 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 20 | fp_O | P13350 | 188 | TMFMTVGIIDRFLQ | HHHHHHHHHHHHCC | c3-AT | multi | 0.014 | 0.011 | 0.091 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 21 | fp_O | P13350 | 189 | MFMTVGIIDRFLQEHP | HHHHHHHHHHHCCCCC | c1aR-4 | multi-selected | 0.016 | 0.016 | 0.102 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 22 | fp_O | P13350 | 208 | NQLQLVGVTAMFLAAK | CCHHHHHHHHHHHHHH | c1aR-4 | multi-selected | 0.048 | 0.014 | 0.075 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 23 | fp_O | P13350 | 208 | NQLQLVGVTAMFLAA | CCHHHHHHHHHHHHH | c1b-4 | multi | 0.049 | 0.014 | 0.075 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 24 | fp_O | P13350 | 209 | QLQLVGVTAMFLAA | CHHHHHHHHHHHHH | c2-5 | multi-selected | 0.049 | 0.012 | 0.069 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 25 | fp_O | P13350 | 209 | QLQLVGVTAMFLAA | CHHHHHHHHHHHHH | c3-AT | multi | 0.049 | 0.012 | 0.069 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 26 | fp_O | P13350 | 240 | DHTYTKAQIRDMEMKI | CCCCCHHHHHHHHHHH | c1a-AT-4 | multi-selected | 0.017 | 0.025 | 0.111 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 27 | fp_O | P13350 | 242 | TYTKAQIRDMEMKI | CCCHHHHHHHHHHH | c2-AT-4 | multi | 0.018 | 0.025 | 0.103 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 28 | fp_O | P13350 | 247 | QIRDMEMKILRVLKFAI | HHHHHHHHHHHHCCCCC | c1c-5 | multi-selected | 0.014 | 0.019 | 0.061 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 29 | fp_O | P13350 | 247 | QIRDMEMKILRVLK | HHHHHHHHHHHHCC | c2-5 | multi-selected | 0.014 | 0.017 | 0.071 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 30 | fp_O | P13350 | 247 | QIRDMEMKILRVLK | HHHHHHHHHHHHCC | c3-4 | multi-selected | 0.014 | 0.017 | 0.071 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 31 | fp_O | P13350 | 249 | RDMEMKILRVLKFAI | HHHHHHHHHHCCCCC | c1b-4 | multi | 0.012 | 0.018 | 0.052 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 32 | fp_O | P13350 | 251 | MEMKILRVLKFAIGR | HHHHHHHHCCCCCCC | c1b-4 | multi | 0.011 | 0.018 | 0.036 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 33 | fp_O | P13350 | 251 | MEMKILRVLKFAIGRPL | HHHHHHHHCCCCCCCCC | c1cR-4 | multi | 0.012 | 0.019 | 0.037 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 34 | fp_O | P13350 | 252 | EMKILRVLKFAIGR | HHHHHHHCCCCCCC | c2-5 | multi-selected | 0.01 | 0.019 | 0.034 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 35 | fp_D | P13350 | 284 | EQHSLAKYLMELVMVD | HHHHHHHHHHHHHHHH | c1a-4 | multi-selected | 0.105 | 0.012 | 0.078 | boundary | MID|Cyclin_N2 superfamily; | 0.0 |
| 36 | fp_D | P13350 | 284 | EQHSLAKYLMELVMVDY | HHHHHHHHHHHHHHHHC | c1c-4 | multi-selected | 0.108 | 0.012 | 0.075 | boundary | MID|Cyclin_N2 superfamily; | 0.0 |
| 37 | fp_D | P13350 | 285 | QHSLAKYLMELVMVD | HHHHHHHHHHHHHHH | c1b-AT-4 | multi | 0.109 | 0.011 | 0.067 | boundary | MID|Cyclin_N2 superfamily; | 0.0 |
| 38 | fp_D | P13350 | 285 | QHSLAKYLMELVMVDY | HHHHHHHHHHHHHHHC | c1a-AT-4 | multi | 0.112 | 0.011 | 0.065 | boundary | MID|Cyclin_N2 superfamily; | 0.0 |
| 39 | fp_D | P13350 | 285 | QHSLAKYLMELVMVDY | HHHHHHHHHHHHHHHC | c1d-AT-4 | multi | 0.112 | 0.011 | 0.065 | boundary | MID|Cyclin_N2 superfamily; | 0.0 |
| 40 | fp_D | P13350 | 291 | YLMELVMVDYDMVHFT | HHHHHHHHHCCCCCCC | c1aR-5 | multi-selected | 0.156 | 0.011 | 0.048 | boundary | MID|Cyclin_N2 superfamily; | 0.0 |
| 41 | fp_D | P13350 | 294 | ELVMVDYDMVHFTP | HHHHHHCCCCCCCH | c3-4 | multi-selected | 0.161 | 0.012 | 0.047 | boundary | MID|Cyclin_N2 superfamily; | 0.0 |
| 42 | fp_D | P13350 | 306 | TPSQIAAASSCLSLKI | CHHHHHHHHHHHHHHH | c1d-AT-4 | uniq | 0.063 | 0.014 | 0.091 | boundary | MID|Cyclin_N2 superfamily; | 0.0 |
| 43 | fp_D | P13350 | 342 | LVPVMQHMAKNIIK | HHHHHHHHHHHHHH | c3-AT | multi | 0.028 | 0.028 | 0.181 | boundary | MID|Cyclin_N2 superfamily; | 0.0 |
| 44 | fp_D | P13350 | 342 | LVPVMQHMAKNIIKVNK | HHHHHHHHHHHHHHHCC | c1c-AT-5 | multi | 0.034 | 0.049 | 0.191 | boundary | MID|Cyclin_N2 superfamily; | 0.0 |
| 45 | fp_D | P13350 | 345 | VMQHMAKNIIKVNK | HHHHHHHHHHHHCC | c3-4 | multi-selected | 0.037 | 0.054 | 0.2 | boundary | MID|Cyclin_N2 superfamily; | 0.0 |
| 46 | fp_D | P13350 | 352 | NIIKVNKGLTKHLTVKN | HHHHHCCCCCCHHHHHH | c1c-5 | uniq | 0.153 | 0.125 | 0.256 | boundary | boundary|Cyclin_N2 superfamily; | 0.0 |
| 47 | fp_D | P13350 | 376 | MKISTIPQLRSDVVVEM | CCCCCCCCCCCHHHHHH | c1c-AT-4 | multi-selected | 0.538 | 0.23 | 0.248 | DISO | boundary|Cyclin_N2 superfamily; | 0.0 |
| 48 | fp_D | P13350 | 377 | KISTIPQLRSDVVVEM | CCCCCCCCCCHHHHHH | c1d-5 | multi | 0.524 | 0.232 | 0.244 | DISO | boundary|Cyclin_N2 superfamily; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment