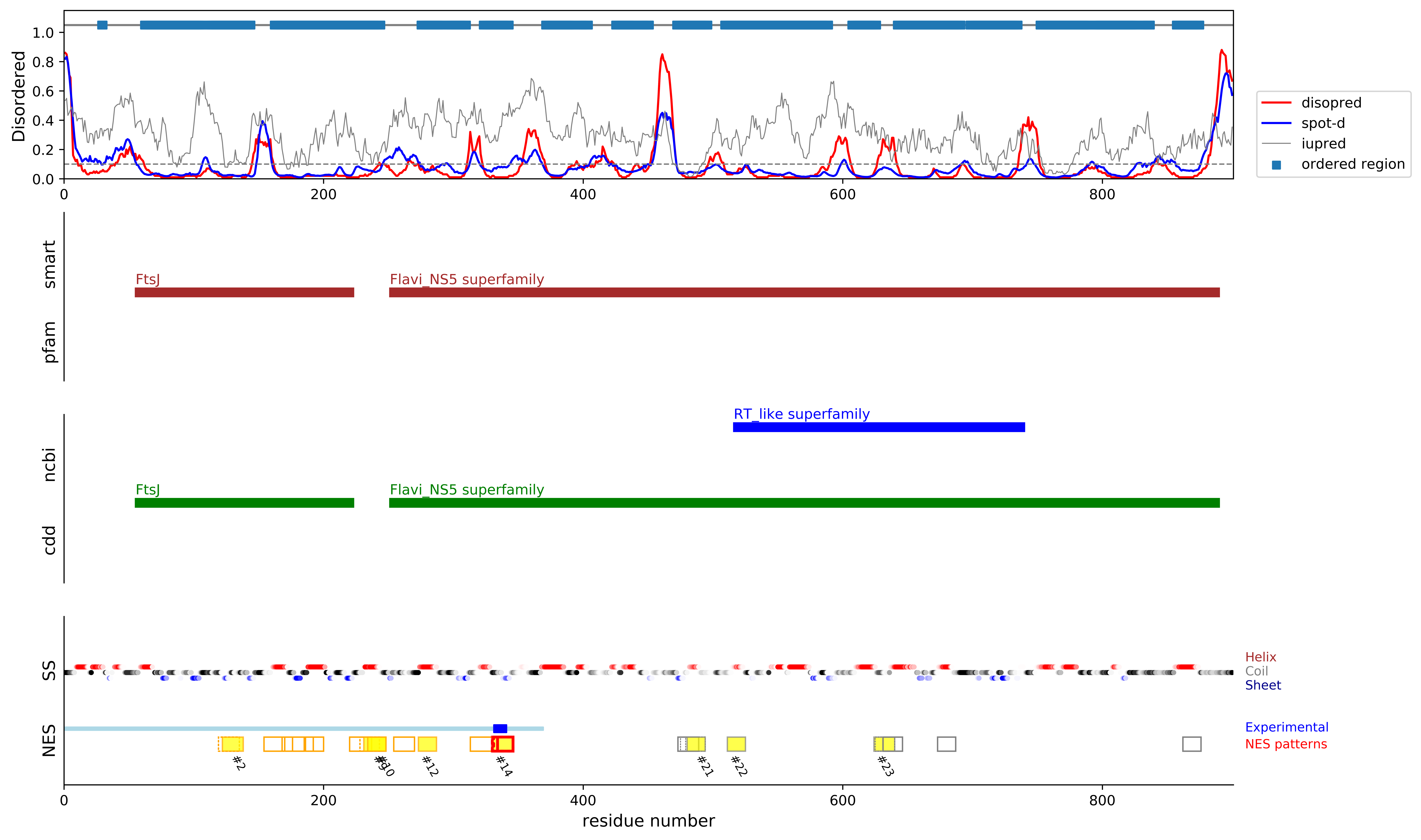

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_beta_D | P14340 | 119 | YGWNLVRLQSGVDVFF ................ | CCCCEEEEECCCCCCC | c1d-4 | multi | 0.015 | 0.025 | 0.165 | boundary | MID|FtsJ; | 0.714 |
| 2 | fp_D | P14340 | 122 | NLVRLQSGVDVFFIPP ................ | CEEEEECCCCCCCCCC | c1a-5 | multi-selected | 0.014 | 0.022 | 0.125 | boundary | MID|FtsJ; | 0.286 |
| 3 | fp_D | P14340 | 154 | PTVEAGRTLRVLNLVE ................ | CCCCCCHHHHHHHHHH | c1a-AT-4 | uniq | 0.121 | 0.137 | 0.326 | boundary | MID|FtsJ; | 0.0 |
| 4 | fp_D | P14340 | 168 | VENWLNNNTQFCIKVLN ................. | HHHHHCCCCCEEEEECC | c1c-AT-4 | uniq | 0.01 | 0.018 | 0.124 | boundary | MID|FtsJ; | 0.25 |
| 5 | fp_O | P14340 | 176 | TQFCIKVLNPYMPSVI ................ | CCEEEEECCCCCHHHH | c1aR-5 | uniq | 0.014 | 0.02 | 0.148 | ORD | MID|FtsJ; | 0.429 |
| 6 | fp_O | P14340 | 186 | YMPSVIEKMEALQR .............. | CCHHHHHHHHHHHH | c3-4 | uniq | 0.02 | 0.024 | 0.227 | ORD | MID|FtsJ; | 0.0 |
| 7 | fp_D | P14340 | 220 | WVSNASGNIVSSVNMIS ................. | EEECCCCCCCCHHHHHH | c1c-AT-5 | uniq | 0.017 | 0.059 | 0.214 | boundary | boundary|FtsJ; boundary|Flavi_NS5 superfamily; | 0.0 |
| 8 | fp_D | P14340 | 228 | IVSSVNMISRMLINR ............... | CCCHHHHHHHHHHHH | c1b-5 | multi | 0.04 | 0.058 | 0.16 | boundary | boundary|FtsJ; boundary|Flavi_NS5 superfamily; | 0.0 |
| 9 | fp_D | P14340 | 231 | SVNMISRMLINRFTMRY ................. | HHHHHHHHHHHHHCCCC | c1c-5 | multi-selected | 0.061 | 0.066 | 0.181 | boundary | boundary|FtsJ; boundary|Flavi_NS5 superfamily; | 0.0 |
| 10 | fp_D | P14340 | 234 | MISRMLINRFTMRY .............. | HHHHHHHHHHCCCC | c2-5 | multi-selected | 0.069 | 0.066 | 0.172 | boundary | boundary|FtsJ; boundary|Flavi_NS5 superfamily; | 0.0 |
| 11 | fp_D | P14340 | 254 | PDVDLGSGTRNIGIES ................ | CCCCCCCCCCCCCCCC | c1a-AT-4 | uniq | 0.066 | 0.167 | 0.427 | boundary | boundary|FtsJ; boundary|Flavi_NS5 superfamily; | 0.0 |
| 12 | fp_D | P14340 | 273 | NLDIIGKRIEKIKQ .............. | CHHHHHHHHHHHHH | c3-4 | uniq | 0.066 | 0.073 | 0.37 | boundary | MID|Flavi_NS5 superfamily; | 0.0 |
| 13 | fp_D | P14340 | 313 | QTGSASSMVNGVVRLLT ................. | CCCCCCCHHHHHHHHHC | c1c-AT-4 | uniq | 0.144 | 0.104 | 0.377 | boundary | MID|Flavi_NS5 superfamily; | 0.0 |
| 14 | cand_D | P14340 | 330 | KPWDVVPMVTQMAMTD ....**..*....... | CCCCCCCCEEEEEECC | c1a-4 | multi-selected | 0.013 | 0.082 | 0.372 | boundary | MID|Flavi_NS5 superfamily; | 0.429 |
| 15 | cand_D | P14340 | 330 | KPWDVVPMVTQMAMTD ....**..*....... | CCCCCCCCEEEEEECC | c1d-4 | multi | 0.013 | 0.082 | 0.372 | boundary | MID|Flavi_NS5 superfamily; | 0.429 |
| 16 | cand_beta_D | P14340 | 331 | PWDVVPMVTQMAMTD ...**..*....... | CCCCCCCEEEEEECC | c1b-4 | multi | 0.013 | 0.084 | 0.38 | boundary | MID|Flavi_NS5 superfamily; | 0.571 |
| 17 | cand_beta_D | P14340 | 334 | VVPMVTQMAMTD **..*....... | CCCCEEEEEECC | c2-rev | multi-selected | 0.012 | 0.087 | 0.406 | boundary | MID|Flavi_NS5 superfamily; | 0.833 |
| 18 | fp_D | P14340 | 473 | AIWYMWLGARFLEFEA | EEEEECCCHHHHHHHH | c1a-AT-5 | multi-selected | 0.021 | 0.042 | 0.038 | __ | MID|Flavi_NS5 superfamily; | 0.143 |
| 19 | fp_D | P14340 | 475 | WYMWLGARFLEFEA | EEECCCHHHHHHHH | c2-AT-4 | multi | 0.016 | 0.041 | 0.035 | boundary | MID|Flavi_NS5 superfamily; | 0.0 |
| 20 | fp_D | P14340 | 479 | LGARFLEFEALGFLN | CCHHHHHHHHHCCCC | c1b-4 | multi | 0.015 | 0.044 | 0.049 | boundary | MID|Flavi_NS5 superfamily; | 0.0 |
| 21 | fp_D | P14340 | 480 | GARFLEFEALGFLN | CHHHHHHHHHCCCC | c2-4 | multi-selected | 0.016 | 0.045 | 0.05 | boundary | MID|Flavi_NS5 superfamily; | 0.0 |
| 22 | fp_D | P14340 | 511 | GLHKLGYILRDVSK | CCCCHHHHHHHHHC | c3-4 | uniq | 0.048 | 0.045 | 0.244 | boundary | MID|Flavi_NS5 superfamily; | 0.0 |
| 23 | fp_D | P14340 | 624 | EGEGVFKSIQHLTITE | HHCCCCCCCCCCCHHH | c1a-4 | multi-selected | 0.186 | 0.06 | 0.266 | boundary | MID|Flavi_NS5 superfamily; | 0.0 |
| 24 | fp_D | P14340 | 625 | GEGVFKSIQHLTITE | HCCCCCCCCCCCHHH | c1b-4 | multi | 0.195 | 0.062 | 0.259 | boundary | MID|Flavi_NS5 superfamily; | 0.0 |
| 25 | fp_D | P14340 | 631 | SIQHLTITEEIAVQN | CCCCCCHHHHHHHHH | c1b-AT-5 | uniq | 0.18 | 0.052 | 0.228 | boundary | MID|Flavi_NS5 superfamily; | 0.0 |
| 26 | fp_D | P14340 | 673 | RFASALTALNDMGK | CHHHHHHHHHHHCC | c3-AT | uniq | 0.013 | 0.047 | 0.247 | boundary | MID|Flavi_NS5 superfamily; | 0.0 |
| 27 | fp_D | P14340 | 862 | NIQAAINQVRSLIG | HHHHHHHHHHHHHC | c3-AT | uniq | 0.016 | 0.059 | 0.265 | boundary | boundary|Flavi_NS5 superfamily; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment