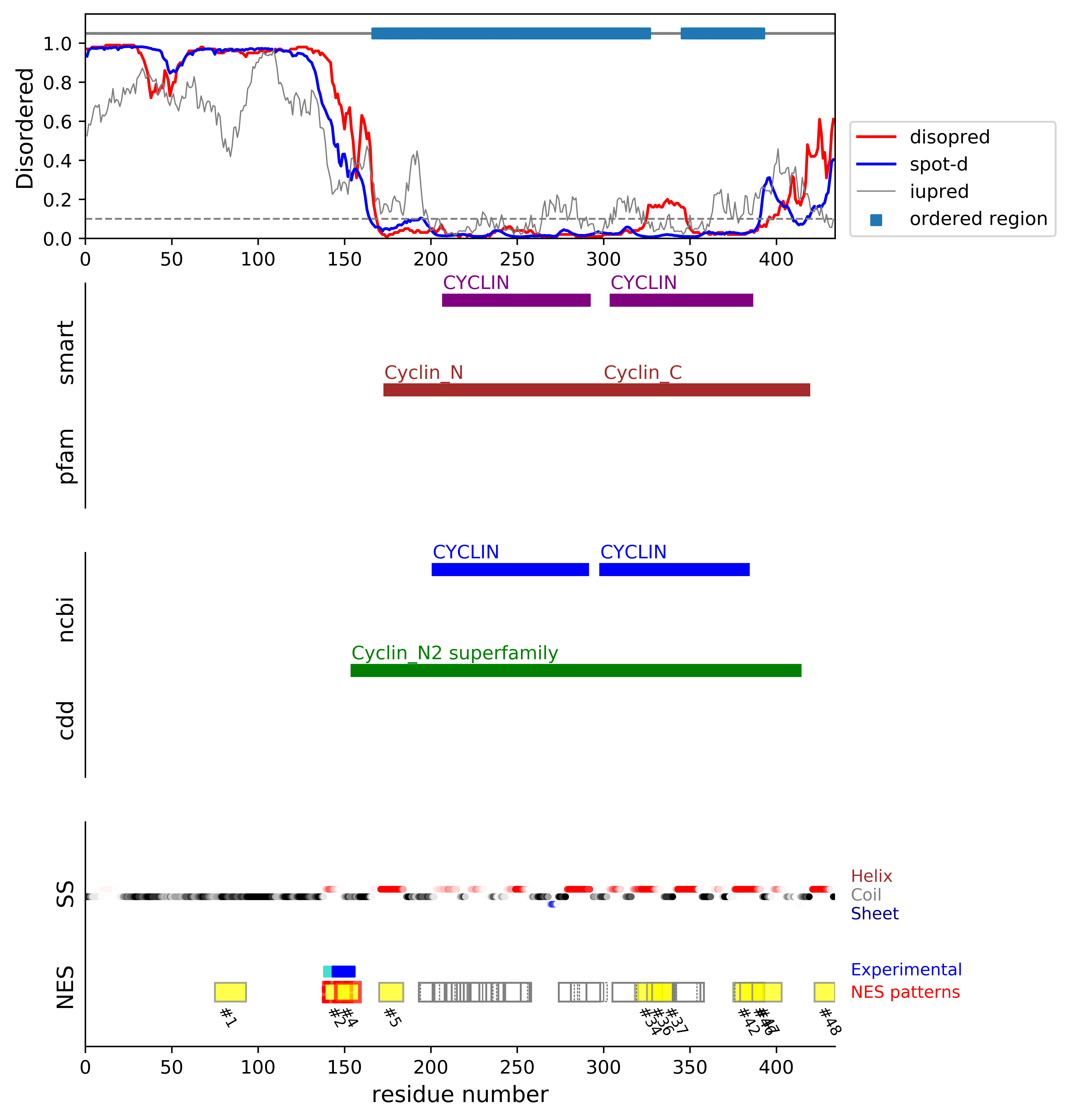

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | P14635 | 75 | IDKKLPKPLEKVPMLVPV | CCCCCCCCCCCCCCCCCC | c4-4 | uniq | 0.957 | 0.964 | 0.557 | DISO | 0.0 | |
| 2 | cand_D | P14635 | 138 | AEEDLCQAFSDVILAV +++++*++*+*+* | CHHHHHHHHHHCCCCC | c1a-4 | multi-selected | 0.759 | 0.491 | 0.31 | __ | boundary|Cyclin_N2 superfamily; | 0.0 |
| 3 | cand_D | P14635 | 138 | AEEDLCQAFSDVILAV +++++*++*+*+* | CHHHHHHHHHHCCCCC | c1d-AT-4 | multi | 0.759 | 0.491 | 0.31 | __ | boundary|Cyclin_N2 superfamily; | 0.0 |
| 4 | cand_D | P14635 | 145 | AFSDVILAVNDVDA +*++*+*+* | HHHHCCCCCCCCCC | c3-4 | uniq | 0.58 | 0.379 | 0.313 | boundary | boundary|Cyclin_N2 superfamily; | 0.0 |
| 5 | fp_D | P14635 | 170 | YVKDIYAYLRQLEE | HHHHHHHHHHHHHH | c3-4 | uniq | 0.026 | 0.054 | 0.169 | boundary | boundary|Cyclin_N2 superfamily; | 0.0 |
| 6 | fp_O | P14635 | 193 | LGREVTGNMRAILIDW | CCCCCCCCCCHHHHHH | c1a-4 | multi-selected | 0.04 | 0.05 | 0.117 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 7 | fp_O | P14635 | 194 | GREVTGNMRAILIDW | CCCCCCCCCHHHHHH | c1b-AT-4 | multi | 0.04 | 0.047 | 0.099 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 8 | fp_O | P14635 | 201 | MRAILIDWLVQVQMKF | CCHHHHHHHHHHHHHC | c1a-4 | multi-selected | 0.033 | 0.017 | 0.044 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 9 | fp_O | P14635 | 202 | RAILIDWLVQVQMKFRL | CHHHHHHHHHHHHHCCC | c1c-4 | multi-selected | 0.029 | 0.016 | 0.043 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 10 | fp_O | P14635 | 202 | RAILIDWLVQVQMKF | CHHHHHHHHHHHHHC | c1b-4 | multi | 0.031 | 0.016 | 0.044 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 11 | fp_O | P14635 | 205 | LIDWLVQVQMKFRLLQ | HHHHHHHHHHHCCCCC | c1d-5 | multi | 0.027 | 0.016 | 0.036 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 12 | fp_O | P14635 | 208 | WLVQVQMKFRLLQE | HHHHHHHHCCCCCH | c3-4 | multi-selected | 0.019 | 0.017 | 0.032 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 13 | fp_O | P14635 | 212 | VQMKFRLLQETMYMTV | HHHHCCCCCHHHHHHH | c1aR-4 | multi-selected | 0.016 | 0.015 | 0.041 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 14 | fp_O | P14635 | 212 | VQMKFRLLQETMYMTV | HHHHCCCCCHHHHHHH | c1d-4 | multi | 0.016 | 0.015 | 0.041 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 15 | fp_O | P14635 | 214 | MKFRLLQETMYMTVSI | HHCCCCCHHHHHHHHH | c1a-AT-4 | multi | 0.014 | 0.015 | 0.046 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 16 | fp_O | P14635 | 215 | KFRLLQETMYMTVSIID | HCCCCCHHHHHHHHHHH | c1c-5 | multi-selected | 0.014 | 0.014 | 0.057 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 17 | fp_O | P14635 | 215 | KFRLLQETMYMTVSI | HCCCCCHHHHHHHHH | c1b-AT-5 | multi | 0.014 | 0.014 | 0.047 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 18 | fp_O | P14635 | 221 | ETMYMTVSIIDRFMQNN | HHHHHHHHHHHCCCCCC | c1cR-4 | multi | 0.015 | 0.016 | 0.073 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 19 | fp_O | P14635 | 222 | TMYMTVSIIDRFMQ | HHHHHHHHHHCCCC | c3-AT | multi | 0.013 | 0.013 | 0.075 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 20 | fp_O | P14635 | 223 | MYMTVSIIDRFMQNNC | HHHHHHHHHCCCCCCC | c1aR-4 | multi-selected | 0.016 | 0.018 | 0.075 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 21 | fp_O | P14635 | 235 | QNNCVPKKMLQLVGVTA | CCCCCCCCCHHHHHHHH | c1c-4 | uniq | 0.042 | 0.023 | 0.06 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 22 | fp_O | P14635 | 242 | KMLQLVGVTAMFIA | CCHHHHHHHHHHHH | c3-AT | multi | 0.045 | 0.012 | 0.044 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 23 | fp_O | P14635 | 242 | KMLQLVGVTAMFIASK | CCHHHHHHHHHHHHHH | c1aR-4 | multi-selected | 0.044 | 0.012 | 0.046 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 24 | fp_O | P14635 | 242 | KMLQLVGVTAMFIAS | CCHHHHHHHHHHHHH | c1b-5 | multi | 0.044 | 0.012 | 0.044 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 25 | fp_O | P14635 | 243 | MLQLVGVTAMFIAS | CHHHHHHHHHHHHH | c2-5 | multi-selected | 0.044 | 0.011 | 0.041 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 26 | fp_O | P14635 | 243 | MLQLVGVTAMFIAS | CHHHHHHHHHHHHH | c3-AT | multi | 0.044 | 0.011 | 0.041 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 27 | fp_O | P14635 | 274 | DNTYTKHQIRQMEMKI | CCCCCHHHHHHHHHHH | c1a-AT-4 | uniq | 0.016 | 0.027 | 0.138 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 28 | fp_O | P14635 | 281 | QIRQMEMKILRALNFGL | HHHHHHHHHHHHCCCCC | c1c-5 | multi-selected | 0.012 | 0.021 | 0.097 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 29 | fp_O | P14635 | 283 | RQMEMKILRALNFGL | HHHHHHHHHHCCCCC | c1b-4 | multi | 0.011 | 0.02 | 0.087 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 30 | fp_O | P14635 | 285 | MEMKILRALNFGLGR | HHHHHHHHCCCCCCC | c1b-AT-4 | multi | 0.01 | 0.022 | 0.064 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 31 | fp_O | P14635 | 285 | MEMKILRALNFGLGRPL | HHHHHHHHCCCCCCCCC | c1cR-4 | multi | 0.011 | 0.022 | 0.066 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 32 | fp_O | P14635 | 286 | EMKILRALNFGLGR | HHHHHHHCCCCCCC | c2-AT-5 | multi | 0.01 | 0.022 | 0.062 | ORD | MID|Cyclin_N2 superfamily; | 0.0 |
| 33 | fp_D | P14635 | 305 | FLRRASKIGEVDVEQ | HHHHHHCCCCCCHHH | c1b-AT-5 | uniq | 0.03 | 0.036 | 0.167 | boundary | MID|Cyclin_N2 superfamily; | 0.0 |
| 34 | fp_D | P14635 | 318 | EQHTLAKYLMELTMLD | HHHHHHHHHHHHHHHH | c1a-4 | multi-selected | 0.112 | 0.012 | 0.091 | boundary | MID|Cyclin_N2 superfamily; | 0.0 |
| 35 | fp_D | P14635 | 319 | QHTLAKYLMELTMLD | HHHHHHHHHHHHHHH | c1b-AT-4 | multi | 0.117 | 0.011 | 0.083 | boundary | MID|Cyclin_N2 superfamily; | 0.0 |
| 36 | fp_D | P14635 | 325 | YLMELTMLDYDMVHFP | HHHHHHHHHCCCCCCC | c1aR-5 | multi-selected | 0.169 | 0.013 | 0.066 | boundary | MID|Cyclin_N2 superfamily; | 0.0 |
| 37 | fp_D | P14635 | 328 | ELTMLDYDMVHFPP | HHHHHHCCCCCCCH | c3-4 | multi-selected | 0.174 | 0.014 | 0.063 | boundary | MID|Cyclin_N2 superfamily; | 0.0 |
| 38 | fp_D | P14635 | 340 | PPSQIAAGAFCLALKI | CHHHHHHHHHHHHHHH | c1a-AT-4 | multi-selected | 0.108 | 0.011 | 0.047 | boundary | MID|Cyclin_N2 superfamily; | 0.0 |
| 39 | fp_D | P14635 | 340 | PPSQIAAGAFCLAL | CHHHHHHHHHHHHH | c2-AT-4 | multi | 0.119 | 0.011 | 0.047 | boundary | MID|Cyclin_N2 superfamily; | 0.0 |
| 40 | fp_D | P14635 | 341 | PSQIAAGAFCLALKILD | HHHHHHHHHHHHHHHHC | c1c-AT-4 | multi-selected | 0.093 | 0.011 | 0.05 | boundary | MID|Cyclin_N2 superfamily; | 0.0 |
| 41 | fp_D | P14635 | 342 | SQIAAGAFCLALKILD | HHHHHHHHHHHHHHHC | c1d-AT-4 | multi | 0.088 | 0.011 | 0.051 | boundary | MID|Cyclin_N2 superfamily; | 0.0 |
| 42 | fp_D | P14635 | 375 | SLLPVMQHLAKNVVMVN | CHHHHHHHHHHHHHHHH | c1c-5 | multi-selected | 0.032 | 0.04 | 0.161 | boundary | MID|Cyclin_N2 superfamily; | 0.0 |
| 43 | fp_D | P14635 | 376 | LLPVMQHLAKNVVMVN | HHHHHHHHHHHHHHHH | c1a-AT-5 | multi | 0.033 | 0.041 | 0.165 | boundary | MID|Cyclin_N2 superfamily; | 0.0 |
| 44 | fp_D | P14635 | 376 | LLPVMQHLAKNVVMVN | HHHHHHHHHHHHHHHH | c1d-5 | multi | 0.033 | 0.041 | 0.165 | boundary | MID|Cyclin_N2 superfamily; | 0.0 |
| 45 | fp_D | P14635 | 376 | LLPVMQHLAKNVVMVNQ | HHHHHHHHHHHHHHHHC | c1c-AT-5 | multi | 0.034 | 0.051 | 0.171 | boundary | boundary|Cyclin_N2 superfamily; | 0.0 |
| 46 | fp_D | P14635 | 379 | VMQHLAKNVVMVNQ | HHHHHHHHHHHHHC | c3-4 | multi-selected | 0.037 | 0.056 | 0.18 | boundary | boundary|Cyclin_N2 superfamily; | 0.0 |
| 47 | fp_D | P14635 | 386 | NVVMVNQGLTKHMTVKN | HHHHHHCCCCCCHHHHH | c1c-5 | uniq | 0.082 | 0.183 | 0.268 | boundary | boundary|Cyclin_N2 superfamily; | 0.0 |
| 48 | fp_D | P14635 | 422 | LVQDLAKAVAKVXX | HHHHHHHHHHCC | c3-4-Ct | uniq | 0.463 | 0.227 | 0.11 | DISO | boundary|Cyclin_N2 superfamily; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment