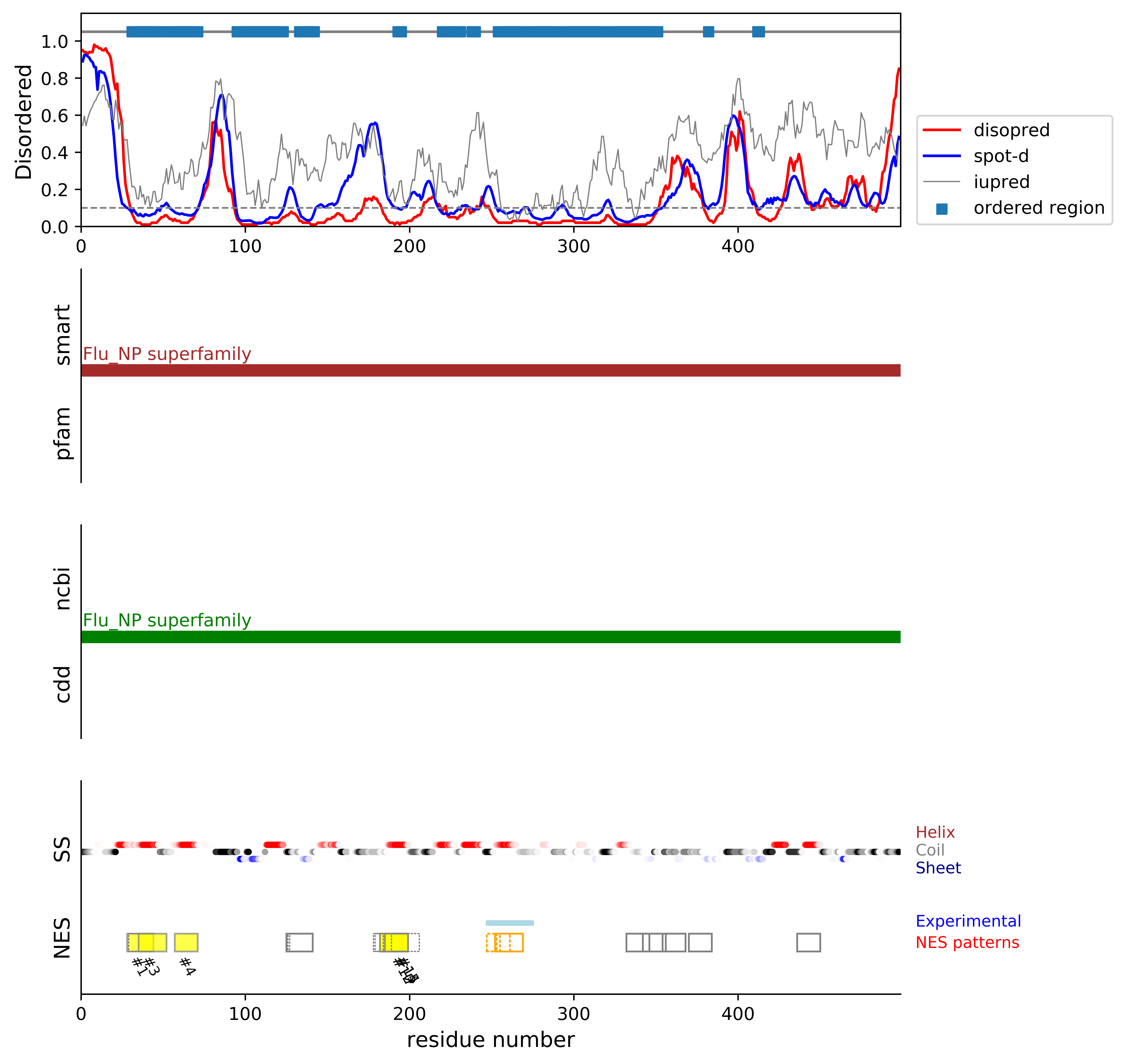

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | P15682 | 28 | SVGKMIGGIGRFYIQM | HHHHHHHHHHHHHHHH | c1a-5 | multi-selected | 0.044 | 0.071 | 0.206 | boundary | MID|Flu_NP superfamily; | 0.0 |
| 2 | fp_D | P15682 | 29 | VGKMIGGIGRFYIQM | HHHHHHHHHHHHHHH | c1b-4 | multi | 0.035 | 0.069 | 0.188 | boundary | MID|Flu_NP superfamily; | 0.0 |
| 3 | fp_D | P15682 | 35 | GIGRFYIQMCTELKLSD | HHHHHHHHHHHHHCCCC | c1c-5 | uniq | 0.024 | 0.075 | 0.163 | boundary | MID|Flu_NP superfamily; | 0.0 |
| 4 | fp_D | P15682 | 57 | IQNSLTIERMVLSA | HHHHHHHHHHHHHH | c2-4 | uniq | 0.034 | 0.067 | 0.273 | boundary | MID|Flu_NP superfamily; | 0.0 |
| 5 | fp_D | P15682 | 125 | NGDDATAGLTHMMIWH | CCCCCCCCCCEEEEEC | c1a-AT-4 | multi-selected | 0.043 | 0.108 | 0.336 | boundary | MID|Flu_NP superfamily; | 0.143 |
| 6 | fp_D | P15682 | 126 | GDDATAGLTHMMIWH | CCCCCCCCCEEEEEC | c1b-AT-4 | multi | 0.042 | 0.104 | 0.331 | boundary | MID|Flu_NP superfamily; | 0.286 |
| 7 | fp_D | P15682 | 127 | DDATAGLTHMMIWH | CCCCCCCCEEEEEC | c2-AT-4 | multi | 0.041 | 0.098 | 0.328 | boundary | MID|Flu_NP superfamily; | 0.286 |
| 8 | fp_D | P15682 | 178 | AAGAAVKGVGTMVMEL | CCCCCCCCCHHHHHHH | c1a-AT-4 | multi | 0.07 | 0.286 | 0.286 | DISO | MID|Flu_NP superfamily; | 0.0 |
| 9 | fp_D | P15682 | 179 | AGAAVKGVGTMVMEL | CCCCCCCCHHHHHHH | c1b-4 | multi | 0.064 | 0.269 | 0.269 | DISO | MID|Flu_NP superfamily; | 0.0 |
| 10 | fp_D | P15682 | 182 | AVKGVGTMVMELIRMIK | CCCCCHHHHHHHHHHHH | c1c-5 | multi-selected | 0.036 | 0.171 | 0.214 | DISO | MID|Flu_NP superfamily; | 0.0 |
| 11 | fp_D | P15682 | 182 | AVKGVGTMVMELIR | CCCCCHHHHHHHHH | c2-AT-5 | multi | 0.04 | 0.186 | 0.231 | DISO | MID|Flu_NP superfamily; | 0.0 |
| 12 | fp_D | P15682 | 182 | AVKGVGTMVMELIR | CCCCCHHHHHHHHH | c3-4 | multi-selected | 0.04 | 0.186 | 0.231 | DISO | MID|Flu_NP superfamily; | 0.0 |
| 13 | fp_D | P15682 | 184 | KGVGTMVMELIRMIK | CCCHHHHHHHHHHHH | c1b-AT-4 | multi | 0.027 | 0.137 | 0.192 | DISO | MID|Flu_NP superfamily; | 0.0 |
| 14 | fp_D | P15682 | 185 | GVGTMVMELIRMIK | CCHHHHHHHHHHHH | c2-5 | multi-selected | 0.024 | 0.124 | 0.186 | DISO | MID|Flu_NP superfamily; | 0.0 |
| 15 | fp_D | P15682 | 185 | GVGTMVMELIRMIK | CCHHHHHHHHHHHH | c3-4 | multi-selected | 0.024 | 0.124 | 0.186 | DISO | MID|Flu_NP superfamily; | 0.0 |
| 16 | fp_D | P15682 | 189 | MVMELIRMIKRGINDRN | HHHHHHHHHHHCCCCCC | c1cR-4 | multi | 0.028 | 0.128 | 0.207 | boundary | MID|Flu_NP superfamily; | 0.0 |
| 17 | fp_D | P15682 | 247 | NPGNAEFEDLIFLA ............. | CCCCCHHHHHHHHH | c2-AT-4 | multi | 0.046 | 0.126 | 0.203 | boundary | MID|Flu_NP superfamily; | 0.0 |
| 18 | fp_D | P15682 | 252 | EFEDLIFLARSALILRG ................. | HHHHHHHHHHHHHHCCC | c1c-AT-5 | multi-selected | 0.025 | 0.082 | 0.102 | boundary | MID|Flu_NP superfamily; | 0.0 |
| 19 | fp_D | P15682 | 253 | FEDLIFLARSALILRG ................ | HHHHHHHHHHHHHCCC | c1d-AT-4 | multi | 0.024 | 0.079 | 0.092 | boundary | MID|Flu_NP superfamily; | 0.0 |
| 20 | fp_D | P15682 | 255 | DLIFLARSALILRG .............. | HHHHHHHHHHHCCC | c3-AT | multi | 0.024 | 0.078 | 0.067 | boundary | MID|Flu_NP superfamily; | 0.0 |
| 21 | fp_D | P15682 | 332 | ACHSAAFEDLRVSS | HHHCCCCCCCCCEE | c2-AT-4 | uniq | 0.011 | 0.042 | 0.143 | boundary | MID|Flu_NP superfamily; | 0.0 |
| 22 | fp_D | P15682 | 342 | RVSSFIRGTKVVPR | CCEEEECCCCCCCC | c3-AT | uniq | 0.071 | 0.075 | 0.272 | boundary | MID|Flu_NP superfamily; | 0.429 |
| 23 | fp_D | P15682 | 354 | PRGKLSTRGVQIAS | CCCCCCCCCEEECC | c2-AT-4 | uniq | 0.289 | 0.184 | 0.472 | boundary | MID|Flu_NP superfamily; | 0.143 |
| 24 | fp_D | P15682 | 370 | NMETMESSTLELRS | CCCCCCCCCCEEEE | c3-AT | uniq | 0.155 | 0.223 | 0.44 | DISO | MID|Flu_NP superfamily; | 0.0 |
| 25 | fp_D | P15682 | 436 | RTSDMRTEIIRLME | CCCCHHHHHHHHHH | c2-AT-4 | uniq | 0.196 | 0.155 | 0.586 | DISO | MID|Flu_NP superfamily; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment