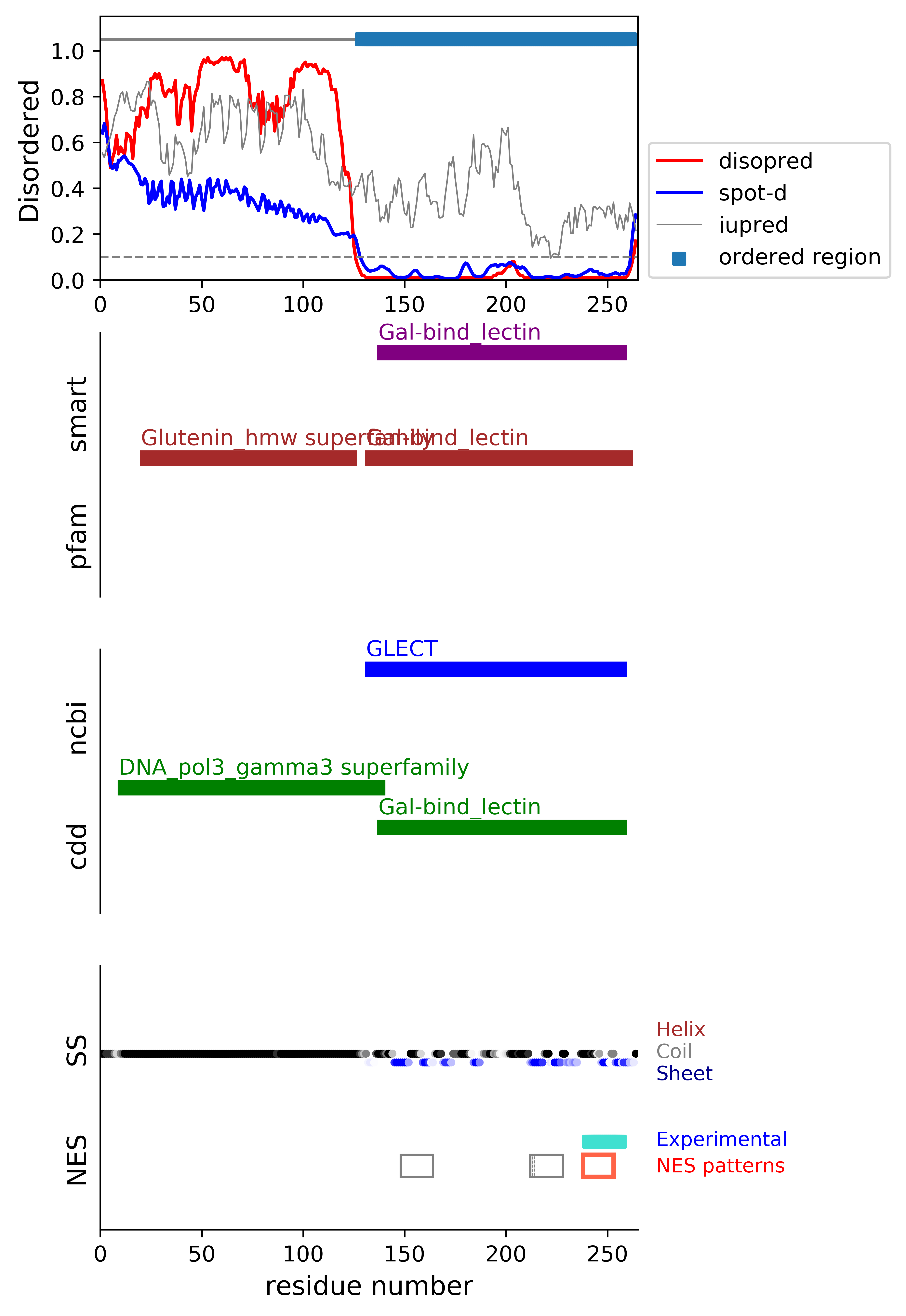

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_O | P16110 | 148 | IMGTVKPNANRIVLDF | EEEEECCCCCCEEEEE | c1a-AT-5 | uniq | 0.01 | 0.019 | 0.351 | ORD | boundary|DNA_pol3_gamma3 superfamily; boundary|Gal-bind_lectin; | 0.143 |
| 2 | fp_beta_O | P16110 | 212 | FKIQVLVEADHFKVAV | EEEEEEECCCEEEEEE | c1a-AT-4 | multi-selected | 0.01 | 0.012 | 0.156 | ORD | MID|Gal-bind_lectin; | 0.571 |
| 3 | fp_beta_O | P16110 | 213 | KIQVLVEADHFKVAV | EEEEEECCCEEEEEE | c1b-AT-5 | multi | 0.01 | 0.012 | 0.151 | ORD | MID|Gal-bind_lectin; | 0.571 |
| 4 | fp_beta_O | P16110 | 214 | IQVLVEADHFKVAV | EEEEECCCEEEEEE | c2-AT-4 | multi | 0.01 | 0.012 | 0.151 | ORD | MID|Gal-bind_lectin; | 0.571 |
| 5 | cand_O | P16110 | 238 | RMKNLREISQLGISG ++++++++++++ | CCCCCCCCCEEEEEC | c1b-5 | uniq | 0.01 | 0.031 | 0.292 | ORD | boundary|Gal-bind_lectin; | 0.286 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment