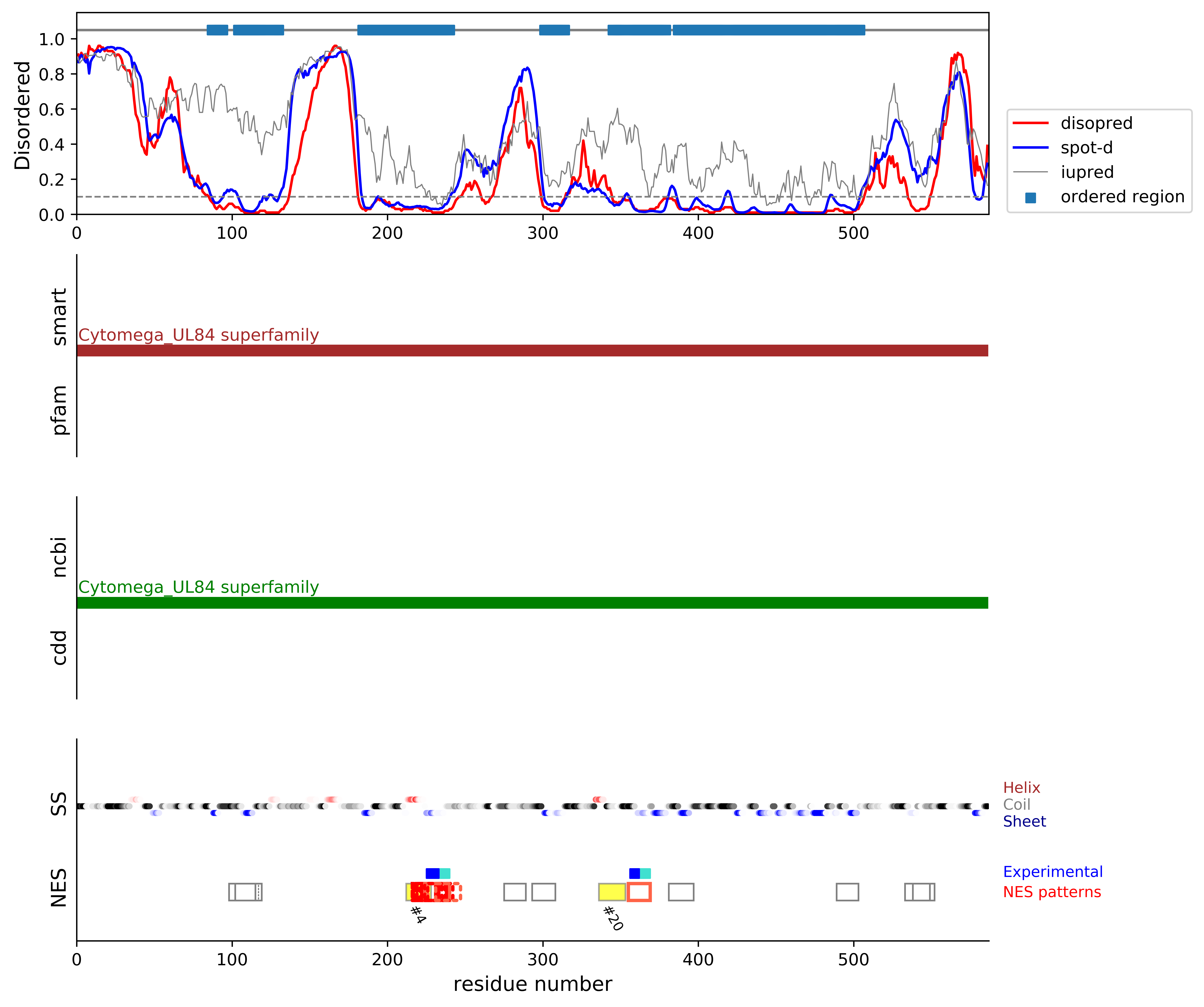

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | P16727 | 98 | RQPRVHRGTYHLIQLHL | CCCCCCCCCEEEEEEEC | c1c-AT-4 | multi-selected | 0.029 | 0.069 | 0.552 | boundary | MID|Cytomega_UL84 superfamily; | 0.375 |
| 2 | fp_beta_D | P16727 | 102 | VHRGTYHLIQLHLDL | CCCCCEEEEEEECCC | c1b-AT-4 | multi | 0.019 | 0.046 | 0.529 | boundary | MID|Cytomega_UL84 superfamily; | 0.857 |
| 3 | fp_beta_D | P16727 | 102 | VHRGTYHLIQLHLDLRP | CCCCCEEEEEEECCCCC | c1c-AT-4 | multi-selected | 0.018 | 0.048 | 0.515 | boundary | MID|Cytomega_UL84 superfamily; | 0.875 |
| 4 | fp_D | P16727 | 212 | LRSVIDQQLTRMAIVR | EHHHHHHHHHHHCEEE | c1a-4 | multi-selected | 0.03 | 0.042 | 0.2 | boundary | MID|Cytomega_UL84 superfamily; | 0.0 |
| 5 | cand_D | P16727 | 216 | IDQQLTRMAIVRLSL *+* | HHHHHHHHCEEEEEE | c1b-4 | multi | 0.027 | 0.04 | 0.146 | boundary | MID|Cytomega_UL84 superfamily; | 0.286 |
| 6 | cand_D | P16727 | 216 | IDQQLTRMAIVRLSLNL *+*++ | HHHHHHHHCEEEEEEEE | c1c-AT-4 | multi | 0.025 | 0.039 | 0.138 | boundary | MID|Cytomega_UL84 superfamily; | 0.375 |
| 7 | cand_D | P16727 | 217 | DQQLTRMAIVRLSL *+* | HHHHHHHCEEEEEE | c2-AT-4 | multi | 0.026 | 0.04 | 0.139 | boundary | MID|Cytomega_UL84 superfamily; | 0.286 |
| 8 | cand_D | P16727 | 217 | DQQLTRMAIVRLSLNL *+*++ | HHHHHHHCEEEEEEEE | c1a-AT-4 | multi | 0.024 | 0.039 | 0.131 | boundary | MID|Cytomega_UL84 superfamily; | 0.429 |
| 9 | cand_beta_D | P16727 | 219 | QLTRMAIVRLSLNLFA *+*++++ | HHHHHCEEEEEEEEEE | c1d-5 | multi | 0.021 | 0.037 | 0.114 | boundary | MID|Cytomega_UL84 superfamily; | 0.714 |
| 10 | cand_beta_D | P16727 | 219 | QLTRMAIVRLSLNL *+*++ | HHHHHCEEEEEEEE | c2-5 | multi | 0.022 | 0.038 | 0.122 | boundary | MID|Cytomega_UL84 superfamily; | 0.571 |
| 11 | cand_beta_D | P16727 | 221 | TRMAIVRLSLNLFALR *+*++++++ | HHHCEEEEEEEEEEEE | c1aR-5 | multi | 0.017 | 0.036 | 0.104 | boundary | MID|Cytomega_UL84 superfamily; | 1.0 |
| 12 | cand_beta_D | P16727 | 222 | RMAIVRLSLNLFALRI *+*+++++++ | HHCEEEEEEEEEEEEC | c1a-5 | multi-selected | 0.016 | 0.037 | 0.099 | boundary | MID|Cytomega_UL84 superfamily; | 1.0 |
| 13 | cand_beta_D | P16727 | 224 | AIVRLSLNLFALRIIT *+*+++++++ | CEEEEEEEEEEEECCC | c1a-5 | multi-selected | 0.016 | 0.04 | 0.092 | boundary | MID|Cytomega_UL84 superfamily; | 1.0 |
| 14 | cand_beta_D | P16727 | 224 | AIVRLSLNLFALRI *+*+++++++ | CEEEEEEEEEEEEC | c2-5 | multi | 0.015 | 0.036 | 0.09 | boundary | MID|Cytomega_UL84 superfamily; | 1.0 |
| 15 | cand_beta_D | P16727 | 226 | VRLSLNLFALRIIT *+*+++++++ | EEEEEEEEEEECCC | c2-4 | multi | 0.014 | 0.039 | 0.088 | boundary | MID|Cytomega_UL84 superfamily; | 1.0 |
| 16 | cand_beta_D | P16727 | 226 | VRLSLNLFALRIITPL *+*+++++++ | EEEEEEEEEEECCCCC | c1aR-4 | multi | 0.016 | 0.048 | 0.094 | boundary | MID|Cytomega_UL84 superfamily; | 1.0 |
| 17 | cand_D | P16727 | 231 | NLFALRIITPLLKRLP +++++++ | EEEEEECCCCCCCCCC | c1aR-5 | multi | 0.034 | 0.084 | 0.141 | boundary | MID|Cytomega_UL84 superfamily; | 0.286 |
| 18 | fp_D | P16727 | 275 | DINNVTENAASVAD | CCCCCCCCCCCCCC | c3-AT | uniq | 0.537 | 0.683 | 0.456 | boundary | MID|Cytomega_UL84 superfamily; | 0.0 |
| 19 | fp_beta_D | P16727 | 293 | TDADLTPTLTVRVRH | CCCCCCCEEEEEEEE | c1b-AT-4 | uniq | 0.141 | 0.28 | 0.326 | boundary | MID|Cytomega_UL84 superfamily; | 0.571 |
| 20 | fp_D | P16727 | 336 | TAKTLGPSVFGRLELDP | HHHHCCCCCCCCCCCCC | c1c-4 | uniq | 0.128 | 0.086 | 0.467 | boundary | MID|Cytomega_UL84 superfamily; | 0.0 |
| 21 | cand_beta_D | P16727 | 355 | SPPDLTLSSLTLYQ *+++++++ | CCCCCEEEEEEEEE | c2-4 | uniq | 0.035 | 0.035 | 0.361 | __ | MID|Cytomega_UL84 superfamily; | 0.714 |
| 22 | fp_beta_D | P16727 | 381 | TEAPADPVAFRLRLRR | CCCCCCCEEEEEEECC | c1d-AT-4 | uniq | 0.049 | 0.074 | 0.307 | boundary | MID|Cytomega_UL84 superfamily; | 0.571 |
| 23 | fp_D | P16727 | 489 | FPRDALLGRLYFIS | ECCCCCCEEEEEEE | c2-AT-4 | uniq | 0.018 | 0.043 | 0.156 | boundary | MID|Cytomega_UL84 superfamily; | 0.429 |
| 24 | fp_beta_D | P16727 | 533 | SQQQLSVLGASIALED | CCCEEEEECCCCCCCC | c1d-4 | multi-selected | 0.056 | 0.279 | 0.316 | DISO | MID|Cytomega_UL84 superfamily; | 0.571 |
| 25 | fp_D | P16727 | 538 | SVLGASIALEDLLP | EEECCCCCCCCCCC | c3-AT | multi-selected | 0.051 | 0.257 | 0.278 | DISO | MID|Cytomega_UL84 superfamily; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment