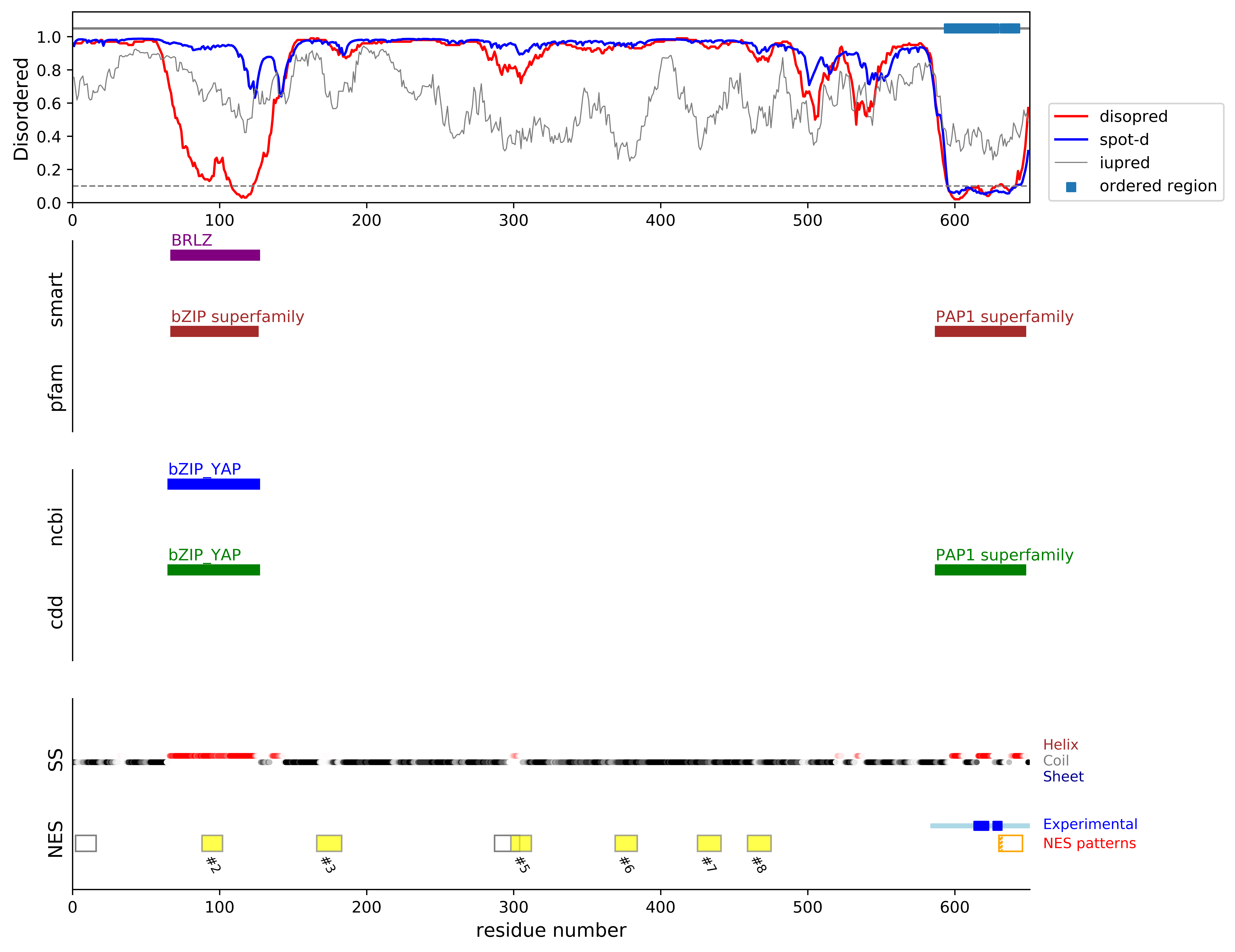

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | P19880 | 2 | SVSTAKRSLDVVSP | CCCCCCCCCCCCCC | c3-AT | uniq | 0.966 | 0.978 | 0.682 | DISO | 0.0 | |
| 2 | fp_D | P19880 | 88 | KMKELEKKVQSLES | HHHHHHHHHHHHHH | c3-4 | uniq | 0.185 | 0.937 | 0.67 | DISO | MID|bZIP_YAP; | 0.0 |
| 3 | fp_D | P19880 | 166 | PNDDIQENVKQKMNFTF | CCCCCHHCCCCCCCCCC | c1c-4 | uniq | 0.952 | 0.961 | 0.7 | DISO | 0.0 | |
| 4 | fp_D | P19880 | 287 | NDFNFENQFDEQVSEFC | CCCCCCCCCCCHHHHHH | c1cR-5 | uniq | 0.817 | 0.946 | 0.402 | DISO | 0.0 | |
| 5 | fp_D | P19880 | 298 | QVSEFCSKMNQVCG | HHHHHHHHCCCCCC | c3-4 | uniq | 0.794 | 0.923 | 0.395 | DISO | 0.0 | |
| 6 | fp_D | P19880 | 369 | YENSFSGFGRLGFDM | CCCCCCCCCCCCCCC | c1b-4 | uniq | 0.943 | 0.953 | 0.311 | DISO | 0.0 | |
| 7 | fp_D | P19880 | 425 | SPFDMNQVTNFFSPGS | CCCCCCCCCCCCCCCC | c1aR-4 | uniq | 0.944 | 0.973 | 0.464 | DISO | 0.0 | |
| 8 | fp_D | P19880 | 459 | SKEDIPFINANLAFPD | CCCCCCCCCCCCCCCC | c1d-4 | uniq | 0.883 | 0.933 | 0.51 | DISO | 0.0 | |
| 9 | fp_D | P19880 | 630 | SERGVVINAEDVQLAL ................ | CCCCCCCCHHHHHHHH | c1a-AT-4 | multi-selected | 0.107 | 0.078 | 0.397 | boundary | boundary|PAP1 superfamily; | 0.0 |
| 10 | fp_D | P19880 | 631 | ERGVVINAEDVQLAL ............... | CCCCCCCHHHHHHHH | c1b-AT-4 | multi | 0.108 | 0.079 | 0.4 | boundary | boundary|PAP1 superfamily; | 0.0 |
| 11 | fp_D | P19880 | 632 | RGVVINAEDVQLAL .............. | CCCCCCHHHHHHHH | c2-AT-4 | multi | 0.108 | 0.08 | 0.4 | boundary | boundary|PAP1 superfamily; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment