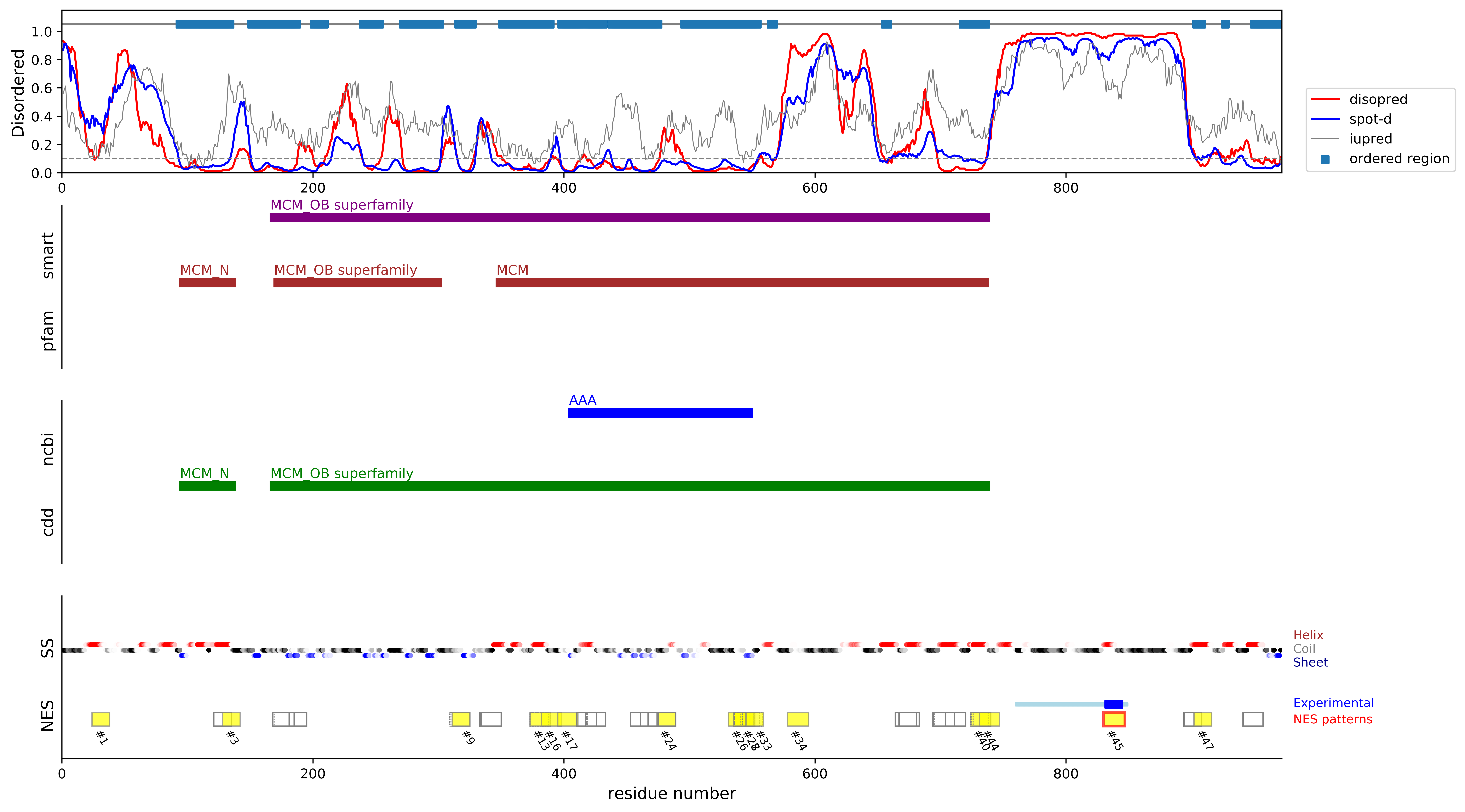

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | P24279 | 24 | RVRRFQEFLDTFTS | HHHHHHHHHHHCCC | c3-4 | uniq | 0.205 | 0.366 | 0.16 | DISO | 0.0 | |
| 2 | fp_D | P24279 | 121 | FIPPAEKALTDLAD | HHHHHHHHHHHHHH | c3-AT | uniq | 0.016 | 0.06 | 0.355 | boundary | small|MCM_N; | 0.0 |
| 3 | fp_D | P24279 | 128 | ALTDLADSMDDVPH | HHHHHHHHCCCCCC | c3-4 | uniq | 0.055 | 0.181 | 0.512 | boundary | 0.0 | |
| 4 | fp_D | P24279 | 168 | PRTLTAQHLNKLVSVEG | CCCCCCCCCCCEEEECC | c1c-AT-4 | multi-selected | 0.019 | 0.028 | 0.322 | boundary | boundary|MCM_OB superfamily; | 0.125 |
| 5 | fp_D | P24279 | 169 | RTLTAQHLNKLVSVEG | CCCCCCCCCCEEEECC | c1d-AT-4 | multi | 0.018 | 0.027 | 0.315 | boundary | boundary|MCM_OB superfamily; | 0.143 |
| 6 | fp_beta_D | P24279 | 181 | SVEGIVTKTSLVRP | EECCEEEECCCCCC | c3-AT | uniq | 0.106 | 0.025 | 0.231 | boundary | boundary|MCM_OB superfamily; | 0.571 |
| 7 | fp_D | P24279 | 309 | SNSNTLIGFKTLILGN | CCCCCCCCCCEEEEEE | c1a-AT-4 | multi | 0.096 | 0.135 | 0.215 | boundary | MID|MCM_OB superfamily; | 0.143 |
| 8 | fp_D | P24279 | 310 | NSNTLIGFKTLILGN | CCCCCCCCCEEEEEE | c1b-4 | multi | 0.085 | 0.114 | 0.202 | boundary | MID|MCM_OB superfamily; | 0.286 |
| 9 | fp_D | P24279 | 311 | SNTLIGFKTLILGN | CCCCCCCCEEEEEE | c2-4 | multi-selected | 0.077 | 0.094 | 0.192 | boundary | MID|MCM_OB superfamily; | 0.286 |
| 10 | fp_D | P24279 | 333 | STGVAARQMLTDFDIRN | CCCCCCCCCCCHHHHHH | c1c-AT-4 | multi-selected | 0.251 | 0.192 | 0.261 | boundary | MID|MCM_OB superfamily; | 0.0 |
| 11 | fp_D | P24279 | 334 | TGVAARQMLTDFDIRN | CCCCCCCCCCHHHHHH | c1a-AT-4 | multi-selected | 0.247 | 0.181 | 0.257 | boundary | MID|MCM_OB superfamily; | 0.0 |
| 12 | fp_D | P24279 | 334 | TGVAARQMLTDFDIRN | CCCCCCCCCCHHHHHH | c1d-AT-4 | multi | 0.247 | 0.181 | 0.257 | boundary | MID|MCM_OB superfamily; | 0.0 |
| 13 | fp_D | P24279 | 373 | GHDHIKKAILLMLMGG | CCHHHHHHHHHHHHCC | c1a-4 | multi-selected | 0.04 | 0.023 | 0.122 | boundary | MID|MCM_OB superfamily; | 0.0 |
| 14 | fp_D | P24279 | 373 | GHDHIKKAILLMLMGG | CCHHHHHHHHHHHHCC | c1d-AT-4 | multi | 0.04 | 0.023 | 0.122 | boundary | MID|MCM_OB superfamily; | 0.0 |
| 15 | fp_D | P24279 | 373 | GHDHIKKAILLMLMG | CCHHHHHHHHHHHHC | c1b-AT-4 | multi | 0.037 | 0.02 | 0.114 | boundary | MID|MCM_OB superfamily; | 0.0 |
| 16 | fp_D | P24279 | 382 | LLMLMGGVEKNLENGS | HHHHHCCCCCCCCCCC | c1aR-4 | uniq | 0.078 | 0.106 | 0.219 | boundary | MID|MCM_OB superfamily; | 0.0 |
| 17 | fp_D | P24279 | 395 | NGSHLRGDINILMVGD | CCCCCCCCCEEEEECC | c1a-4 | uniq | 0.036 | 0.044 | 0.309 | boundary | MID|MCM_OB superfamily; | 0.286 |
| 18 | fp_D | P24279 | 410 | DPSTAKSQLLRFVLNT | CCCHHHHHHHHHHHHH | c1a-AT-4 | uniq | 0.084 | 0.04 | 0.155 | boundary | MID|MCM_OB superfamily; | 0.0 |
| 19 | fp_D | P24279 | 417 | QLLRFVLNTASLAIAT | HHHHHHHHHCCCEECC | c1a-AT-5 | multi-selected | 0.066 | 0.053 | 0.164 | boundary | MID|MCM_OB superfamily; | 0.0 |
| 20 | fp_D | P24279 | 418 | LLRFVLNTASLAIAT | HHHHHHHHCCCEECC | c1b-AT-5 | multi | 0.062 | 0.054 | 0.159 | boundary | MID|MCM_OB superfamily; | 0.0 |
| 21 | fp_D | P24279 | 419 | LRFVLNTASLAIAT | HHHHHHHCCCEECC | c2-AT-4 | multi | 0.059 | 0.055 | 0.157 | boundary | MID|MCM_OB superfamily; | 0.0 |
| 22 | fp_beta_D | P24279 | 453 | GERRLEAGAMVLAD | CCEEECCCEEEEEC | c2-AT-4 | uniq | 0.023 | 0.026 | 0.315 | boundary | MID|MCM_OB superfamily; | 0.571 |
| 23 | fp_beta_D | P24279 | 461 | AMVLADRGVVCIDE | EEEEECCCEEEECC | c3-AT | uniq | 0.021 | 0.016 | 0.147 | boundary | MID|MCM_OB superfamily; | 0.571 |
| 24 | fp_D | P24279 | 474 | EFDKMTDVDRVAIHE | CCCCCCCCCHHHHHH | c1b-5 | multi-selected | 0.219 | 0.067 | 0.26 | boundary | MID|MCM_OB superfamily; | 0.0 |
| 25 | fp_D | P24279 | 475 | FDKMTDVDRVAIHE | CCCCCCCCHHHHHH | c2-AT-4 | multi-selected | 0.229 | 0.07 | 0.268 | boundary | MID|MCM_OB superfamily; | 0.0 |
| 26 | fp_D | P24279 | 531 | QNIALPDSLLSRFDLLF | CCCCCCCCCCCCCEEEE | c1c-4 | multi-selected | 0.024 | 0.04 | 0.236 | boundary | MID|MCM_OB superfamily; | 0.0 |
| 27 | fp_D | P24279 | 535 | LPDSLLSRFDLLFVVTD | CCCCCCCCCEEEEEEEC | c1c-4 | multi-selected | 0.026 | 0.029 | 0.166 | boundary | MID|MCM_OB superfamily; | 0.375 |
| 28 | fp_D | P24279 | 535 | LPDSLLSRFDLLFVVT | CCCCCCCCCEEEEEEE | c1a-4 | multi-selected | 0.024 | 0.029 | 0.167 | boundary | MID|MCM_OB superfamily; | 0.286 |
| 29 | fp_D | P24279 | 536 | PDSLLSRFDLLFVVTD | CCCCCCCCEEEEEEEC | c1d-4 | multi | 0.026 | 0.027 | 0.154 | boundary | MID|MCM_OB superfamily; | 0.429 |
| 30 | fp_D | P24279 | 536 | PDSLLSRFDLLFVVT | CCCCCCCCEEEEEEE | c1b-4 | multi | 0.024 | 0.027 | 0.154 | boundary | MID|MCM_OB superfamily; | 0.429 |
| 31 | fp_beta_D | P24279 | 541 | SRFDLLFVVTDDINEIR | CCCEEEEEEECCCCHHH | c1cR-5 | multi | 0.049 | 0.048 | 0.158 | boundary | MID|MCM_OB superfamily; | 0.75 |
| 32 | fp_beta_D | P24279 | 542 | RFDLLFVVTDDINE | CCEEEEEEECCCCH | c3-AT | multi | 0.041 | 0.038 | 0.15 | boundary | MID|MCM_OB superfamily; | 0.857 |
| 33 | fp_D | P24279 | 545 | LLFVVTDDINEIRD | EEEEEECCCCHHHH | c3-4 | multi-selected | 0.061 | 0.061 | 0.187 | boundary | MID|MCM_OB superfamily; | 0.429 |
| 34 | fp_D | P24279 | 578 | EGEPVRERLNLSLAVGE | CCCCHHHCCCCCCCCCC | c1c-4 | uniq | 0.841 | 0.521 | 0.403 | DISO | MID|MCM_OB superfamily; | 0.0 |
| 35 | fp_D | P24279 | 664 | KERVIPQLTQEAINVIV | HHCCCCCCCHHHHHHHH | c1c-AT-4 | multi-selected | 0.174 | 0.124 | 0.319 | DISO | MID|MCM_OB superfamily; | 0.0 |
| 36 | fp_D | P24279 | 667 | VIPQLTQEAINVIVKN | CCCCCCHHHHHHHHHH | c1a-AT-5 | multi-selected | 0.197 | 0.125 | 0.344 | DISO | MID|MCM_OB superfamily; | 0.0 |
| 37 | fp_D | P24279 | 694 | KSPITARTLETLIRLAT | CCCCCHHHHHHHHHHHH | c1c-AT-4 | multi-selected | 0.071 | 0.151 | 0.474 | boundary | MID|MCM_OB superfamily; | 0.0 |
| 38 | fp_D | P24279 | 695 | SPITARTLETLIRLAT | CCCCHHHHHHHHHHHH | c1d-AT-4 | multi | 0.056 | 0.144 | 0.457 | boundary | MID|MCM_OB superfamily; | 0.0 |
| 39 | fp_D | P24279 | 704 | TLIRLATAHAKVRLSK | HHHHHHHHHHHHHCCC | c1d-AT-5 | uniq | 0.024 | 0.113 | 0.376 | boundary | boundary|MCM_OB superfamily; | 0.0 |
| 40 | fp_D | P24279 | 724 | VDAKVAANLLRFALLG | HHHHHHHHHHHHHHCC | c1a-4 | multi-selected | 0.061 | 0.095 | 0.276 | boundary | boundary|MCM_OB superfamily; | 0.0 |
| 41 | fp_D | P24279 | 724 | VDAKVAANLLRFAL | HHHHHHHHHHHHHH | c2-AT-4 | multi | 0.039 | 0.083 | 0.271 | boundary | boundary|MCM_OB superfamily; | 0.0 |
| 42 | fp_D | P24279 | 725 | DAKVAANLLRFALLG | HHHHHHHHHHHHHCC | c1b-AT-4 | multi | 0.064 | 0.095 | 0.271 | boundary | boundary|MCM_OB superfamily; | 0.0 |
| 43 | fp_D | P24279 | 726 | AKVAANLLRFALLG | HHHHHHHHHHHHCC | c2-AT-4 | multi | 0.067 | 0.096 | 0.27 | boundary | boundary|MCM_OB superfamily; | 0.0 |
| 44 | fp_D | P24279 | 731 | NLLRFALLGEDIGNDI | HHHHHHHCCCCCCCCH | c1aR-4 | multi-selected | 0.269 | 0.268 | 0.335 | boundary | boundary|MCM_OB superfamily; | 0.0 |
| 45 | cand_D | P24279 | 830 | EEAELQRRLQLGLRVSP ....*...*.*.*.... | HHHHHHHHHHCCCCCCC | c1c-4 | uniq | 0.972 | 0.873 | 0.607 | DISO | 0.0 | |
| 46 | fp_D | P24279 | 894 | EPGTISTGRLSLIS | CCCCCCHHHHHHHH | c2-AT-4 | uniq | 0.274 | 0.231 | 0.366 | DISO | 0.0 | |
| 47 | fp_D | P24279 | 902 | RLSLISGIIARLMQ | HHHHHHHHHHHHHH | c3-4 | uniq | 0.079 | 0.11 | 0.277 | DISO | 0.0 | |
| 48 | fp_D | P24279 | 941 | EKFSAQEYLAGLKIMS | CCCCHHHHHHHHHHHH | c1a-AT-4 | uniq | 0.132 | 0.05 | 0.369 | boundary | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment