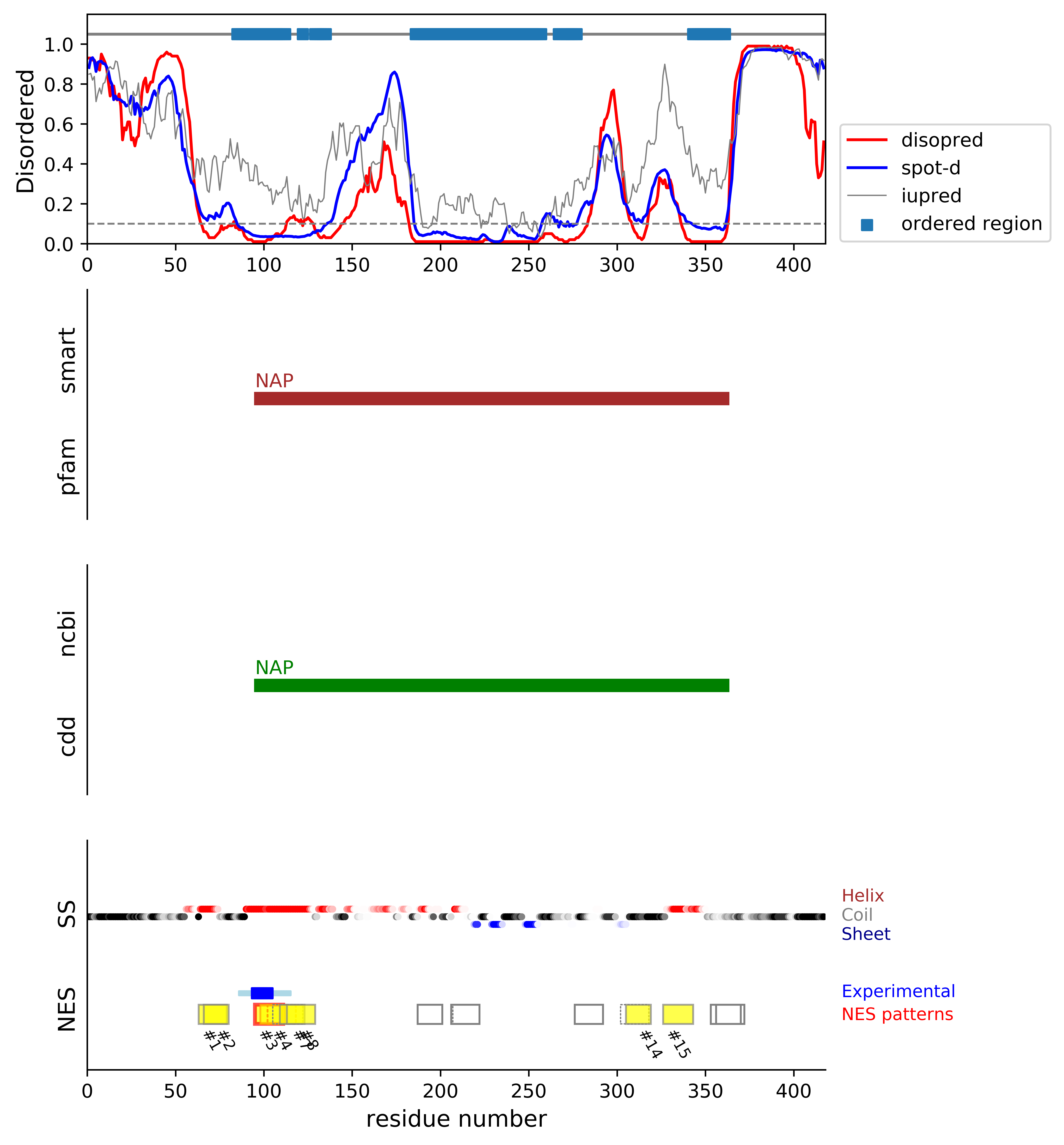

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | P25293 | 63 | QPLLLQSIQDRLGSLV | CHHHHHHHHHHHCCCC | c1aR-5 | multi-selected | 0.064 | 0.148 | 0.368 | boundary | boundary|NAP; | 0.0 |
| 2 | fp_D | P25293 | 66 | LLQSIQDRLGSLVG | HHHHHHHHHCCCCC | c3-4 | multi-selected | 0.054 | 0.148 | 0.367 | boundary | boundary|NAP; | 0.0 |
| 3 | cand_D | P25293 | 95 | KLLSLKTLQSELFEVE .**.*..*........ | HHHHHHHHHHHHHHHH | c1aR-5 | multi-selected | 0.024 | 0.037 | 0.284 | boundary | boundary|NAP; | 0.0 |
| 4 | fp_D | P25293 | 98 | SLKTLQSELFEVEK .*..*......... | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.03 | 0.036 | 0.267 | boundary | boundary|NAP; | 0.0 |
| 5 | fp_D | P25293 | 102 | LQSELFEVEKEFQVEM *............ | HHHHHHHHHHHHHHHH | c1d-4 | multi | 0.068 | 0.036 | 0.247 | boundary | boundary|NAP; | 0.0 |
| 6 | fp_D | P25293 | 105 | ELFEVEKEFQVEMFELE .......... | HHHHHHHHHHHHHHHHH | c1cR-5 | multi | 0.088 | 0.036 | 0.237 | boundary | boundary|NAP; | 0.0 |
| 7 | fp_D | P25293 | 109 | VEKEFQVEMFELEN ...... | HHHHHHHHHHHHHH | c2-4 | multi-selected | 0.101 | 0.036 | 0.231 | boundary | boundary|NAP; | 0.0 |
| 8 | fp_D | P25293 | 113 | FQVEMFELENKFLQKY .. | HHHHHHHHHHHHHHHH | c1aR-4 | multi-selected | 0.118 | 0.039 | 0.207 | boundary | boundary|NAP; | 0.0 |
| 9 | fp_D | P25293 | 187 | SFWLTALENLPIVC | CHHHHHHHCCHHHC | c2-AT-5 | uniq | 0.01 | 0.05 | 0.139 | boundary | MID|NAP; | 0.0 |
| 10 | fp_O | P25293 | 206 | RDAEVLEYLQDIGLEY | CHHHHHHHHCCCCEEE | c1a-4 | multi-selected | 0.01 | 0.029 | 0.195 | ORD | MID|NAP; | 0.0 |
| 11 | fp_O | P25293 | 207 | DAEVLEYLQDIGLEY | HHHHHHHHCCCCEEE | c1b-4 | multi | 0.01 | 0.028 | 0.196 | ORD | MID|NAP; | 0.0 |
| 12 | fp_D | P25293 | 276 | WKDNAHNVTVDLEMRK | CCCCCCCCCCCCHHHC | c1d-AT-4 | uniq | 0.176 | 0.223 | 0.365 | boundary | MID|NAP; | 0.0 |
| 13 | fp_D | P25293 | 302 | TIEKITPIESFFNFFD | EEEECCCCCCCCCCCC | c1d-5 | multi | 0.134 | 0.16 | 0.322 | DISO | MID|NAP; | 0.0 |
| 14 | fp_D | P25293 | 305 | KITPIESFFNFFDP | ECCCCCCCCCCCCC | c3-4 | multi-selected | 0.086 | 0.145 | 0.324 | DISO | MID|NAP; | 0.0 |
| 15 | fp_D | P25293 | 326 | QDEELEEDLEERLALDY | CCHHHHHHHHHHHHHHH | c1c-4 | uniq | 0.171 | 0.212 | 0.633 | boundary | boundary|NAP; | 0.0 |
| 16 | fp_D | P25293 | 353 | IPRAVDWFTGAALEFEF | CCCCCCCCCCCCCCCCC | c1c-AT-4 | multi-selected | 0.262 | 0.239 | 0.422 | boundary | boundary|NAP; | 0.0 |
| 17 | fp_D | P25293 | 356 | AVDWFTGAALEFEFEE | CCCCCCCCCCCCCCCC | c1a-AT-5 | multi-selected | 0.394 | 0.35 | 0.482 | boundary | boundary|NAP; | 0.0 |
| 18 | fp_D | P25293 | 356 | AVDWFTGAALEFEFEE | CCCCCCCCCCCCCCCC | c1d-AT-5 | multi | 0.394 | 0.35 | 0.482 | boundary | boundary|NAP; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment