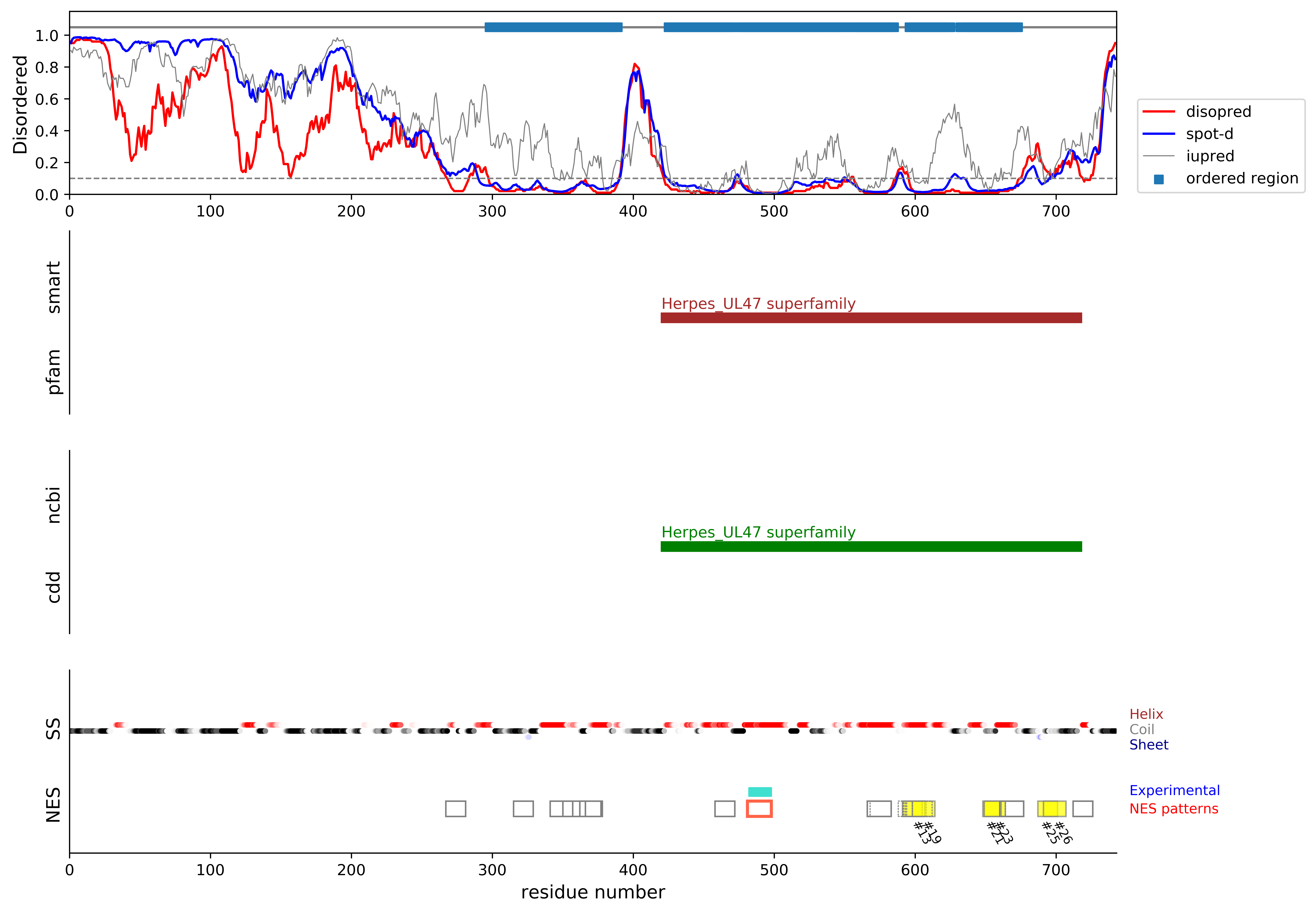

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | P30021 | 267 | VRPSAFFAQMPLDA | CCHHHHHHHCCCCC | c2-AT-4 | uniq | 0.042 | 0.138 | 0.366 | DISO | 0.0 | |
| 2 | fp_O | P30021 | 315 | VDPEFSIGGMYVGA | CCCCCCCCCEEECC | c2-4 | uniq | 0.028 | 0.04 | 0.297 | ORD | 0.143 | |
| 3 | fp_O | P30021 | 341 | AMKQAMALQYRLGVGG | HHHHHHHHHHHHHHHH | c1d-AT-5 | uniq | 0.013 | 0.021 | 0.126 | ORD | 0.0 | |
| 4 | fp_O | P30021 | 350 | YRLGVGGLCRAVDGAR | HHHHHHHHHHHHHCCC | c1aR-4 | uniq | 0.037 | 0.044 | 0.182 | ORD | 0.0 | |
| 5 | fp_D | P30021 | 362 | DGARMPPTEALLFLAA | HCCCCCCHHHHHHHHH | c1d-AT-4 | multi-selected | 0.046 | 0.054 | 0.197 | boundary | 0.0 | |
| 6 | fp_D | P30021 | 362 | DGARMPPTEALLFLA | HCCCCCCHHHHHHHH | c1b-AT-4 | multi-selected | 0.048 | 0.056 | 0.194 | boundary | 0.0 | |
| 7 | fp_O | P30021 | 458 | GLQSAELYLLALRH | HHHHHHHHHHHHCC | c2-AT-5 | multi-selected | 0.022 | 0.026 | 0.075 | ORD | MID|Herpes_UL47 superfamily; | 0.0 |
| 8 | fp_O | P30021 | 458 | GLQSAELYLLALRH | HHHHHHHHHHHHCC | c3-AT | multi-selected | 0.022 | 0.026 | 0.075 | ORD | MID|Herpes_UL47 superfamily; | 0.0 |
| 9 | cand_O | P30021 | 481 | ERYALSAYLTLFVALAE +++++++++++ | HHHHHHHHHHHHHHHHH | c1c-4 | uniq | 0.015 | 0.011 | 0.028 | ORD | MID|Herpes_UL47 superfamily; | 0.0 |
| 10 | fp_D | P30021 | 566 | GEAYVAVRTATTLLMAE | HHHHHHHHHHHHHHHHH | c1c-AT-4 | multi-selected | 0.012 | 0.017 | 0.087 | boundary | MID|Herpes_UL47 superfamily; | 0.0 |
| 11 | fp_D | P30021 | 568 | AYVAVRTATTLLMAE | HHHHHHHHHHHHHHH | c1b-AT-4 | multi | 0.012 | 0.017 | 0.093 | boundary | MID|Herpes_UL47 superfamily; | 0.0 |
| 12 | fp_D | P30021 | 588 | ERRDVREMTAAFLGVGL | CCCCHHHHHHHHHHHHH | c1c-AT-4 | multi | 0.081 | 0.053 | 0.137 | boundary | MID|Herpes_UL47 superfamily; | 0.0 |
| 13 | fp_D | P30021 | 591 | DVREMTAAFLGVGLIA | CHHHHHHHHHHHHHHH | c1a-5 | multi-selected | 0.058 | 0.034 | 0.122 | boundary | MID|Herpes_UL47 superfamily; | 0.0 |
| 14 | fp_D | P30021 | 591 | DVREMTAAFLGVGLIA | CHHHHHHHHHHHHHHH | c1d-AT-5 | multi | 0.058 | 0.034 | 0.122 | boundary | MID|Herpes_UL47 superfamily; | 0.0 |
| 15 | fp_D | P30021 | 591 | DVREMTAAFLGVGL | CHHHHHHHHHHHHH | c2-AT-5 | multi | 0.065 | 0.036 | 0.128 | __ | MID|Herpes_UL47 superfamily; | 0.0 |
| 16 | fp_D | P30021 | 592 | VREMTAAFLGVGLIA | HHHHHHHHHHHHHHH | c1b-AT-4 | multi | 0.051 | 0.028 | 0.116 | __ | MID|Herpes_UL47 superfamily; | 0.0 |
| 17 | fp_D | P30021 | 593 | REMTAAFLGVGLIA | HHHHHHHHHHHHHH | c2-AT-4 | multi | 0.044 | 0.024 | 0.112 | boundary | MID|Herpes_UL47 superfamily; | 0.0 |
| 18 | fp_D | P30021 | 594 | EMTAAFLGVGLIAQ | HHHHHHHHHHHHHH | c3-AT | multi | 0.036 | 0.021 | 0.104 | boundary | MID|Herpes_UL47 superfamily; | 0.0 |
| 19 | fp_D | P30021 | 598 | AFLGVGLIAQRLMGSL | HHHHHHHHHHHHHHHH | c1aR-4 | multi-selected | 0.012 | 0.017 | 0.113 | boundary | MID|Herpes_UL47 superfamily; | 0.0 |
| 20 | fp_D | P30021 | 598 | AFLGVGLIAQRLMG | HHHHHHHHHHHHHH | c3-AT | multi | 0.012 | 0.016 | 0.097 | boundary | MID|Herpes_UL47 superfamily; | 0.0 |
| 21 | fp_D | P30021 | 648 | AALPLVRPVSLVEFWE | HHCCCCCCCCHHHHHH | c1a-4 | multi-selected | 0.012 | 0.024 | 0.081 | boundary | MID|Herpes_UL47 superfamily; | 0.0 |
| 22 | fp_D | P30021 | 649 | ALPLVRPVSLVEFWE | HCCCCCCCCHHHHHH | c1b-5 | multi | 0.013 | 0.025 | 0.082 | boundary | MID|Herpes_UL47 superfamily; | 0.0 |
| 23 | fp_D | P30021 | 649 | ALPLVRPVSLVE | HCCCCCCCCHHH | c2-rev | multi-selected | 0.011 | 0.024 | 0.07 | boundary | MID|Herpes_UL47 superfamily; | 0.0 |
| 24 | fp_D | P30021 | 660 | EFWEARDGVMRELRLRP | HHHHHHHHHHHHCCCCC | c1c-AT-5 | uniq | 0.046 | 0.043 | 0.226 | boundary | MID|Herpes_UL47 superfamily; | 0.0 |
| 25 | fp_D | P30021 | 687 | RVMELYLSLDSIEA | CEEEECCCCCCCCC | c3-4 | multi-selected | 0.171 | 0.094 | 0.156 | boundary | boundary|Herpes_UL47 superfamily; | 0.286 |
| 26 | fp_D | P30021 | 691 | LYLSLDSIEALVGREP | ECCCCCCCCCCCCCCC | c1aR-4 | multi-selected | 0.154 | 0.119 | 0.185 | DISO | boundary|Herpes_UL47 superfamily; | 0.0 |
| 27 | fp_D | P30021 | 712 | VLGPLVDIAEALAD | CCCCHHHHHHHHHC | c3-AT | uniq | 0.141 | 0.223 | 0.312 | DISO | boundary|Herpes_UL47 superfamily; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment