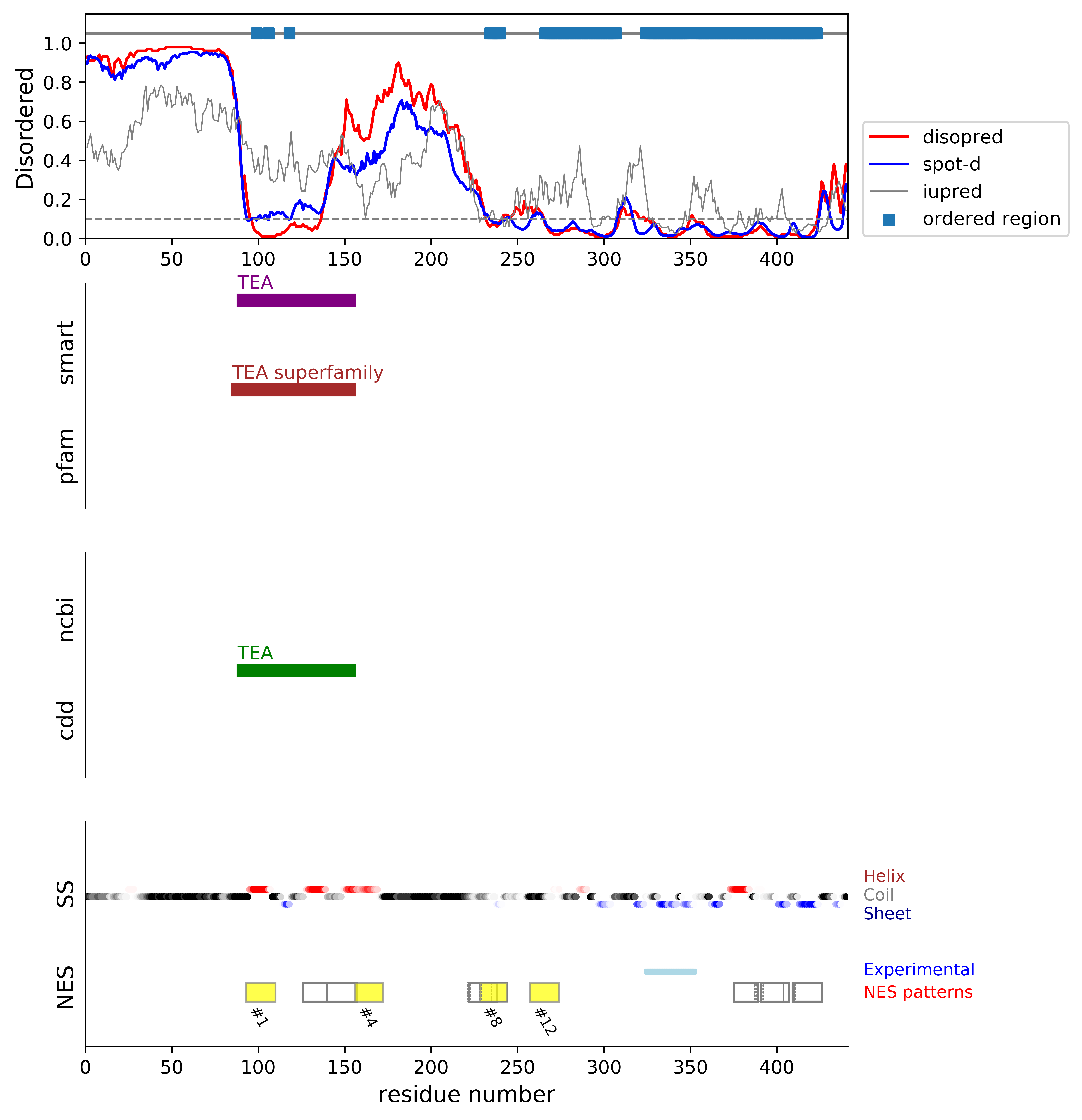

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | P30052 | 93 | WSPDIEQSFQEALSIYP | CCHHHHHHHHHHHHHCC | c1c-4 | uniq | 0.052 | 0.109 | 0.39 | DISO | boundary|TEA; | 0.0 |
| 2 | fp_D | P30052 | 126 | GRNELIARYIKLRT | CHHHHHHHHHHHHH | c2-AT-4 | uniq | 0.085 | 0.156 | 0.353 | DISO | boundary|TEA; | 0.0 |
| 3 | fp_D | P30052 | 140 | GKTRTRKQVSSHIQVLA | CCCCCCCCCHHHHHHHH | c1c-AT-4 | uniq | 0.483 | 0.364 | 0.426 | DISO | boundary|TEA; | 0.0 |
| 4 | fp_D | P30052 | 156 | ARRKLREIQAKIKVQF | HHHHHHHHHHHHHHCC | c1d-4 | uniq | 0.585 | 0.388 | 0.253 | DISO | boundary|TEA; | 0.0 |
| 5 | fp_D | P30052 | 221 | EGRAIATHKFRLLE | CCCCCCCCCCCCCC | c2-AT-4 | multi | 0.224 | 0.192 | 0.179 | __ | 0.0 | |
| 6 | fp_D | P30052 | 222 | GRAIATHKFRLLEFTA | CCCCCCCCCCCCCCCC | c1a-AT-4 | multi-selected | 0.185 | 0.168 | 0.156 | boundary | 0.0 | |
| 7 | fp_D | P30052 | 223 | RAIATHKFRLLEFTA | CCCCCCCCCCCCCCC | c1b-AT-4 | multi | 0.175 | 0.163 | 0.146 | boundary | 0.0 | |
| 8 | fp_D | P30052 | 228 | HKFRLLEFTAFMEIQR | CCCCCCCCCCEEEECC | c1aR-4 | multi-selected | 0.109 | 0.109 | 0.102 | boundary | 0.143 | |
| 9 | fp_D | P30052 | 228 | HKFRLLEFTAFMEIQR | CCCCCCCCCCEEEECC | c1a-AT-4 | multi | 0.109 | 0.109 | 0.102 | boundary | 0.143 | |
| 10 | fp_D | P30052 | 228 | HKFRLLEFTAFMEIQR | CCCCCCCCCCEEEECC | c1d-4 | multi | 0.109 | 0.109 | 0.102 | boundary | 0.143 | |
| 11 | fp_D | P30052 | 229 | KFRLLEFTAFMEIQR | CCCCCCCCCEEEECC | c1b-AT-5 | multi | 0.099 | 0.103 | 0.104 | boundary | 0.286 | |
| 12 | fp_D | P30052 | 257 | GKPSFSDPLLETVDIRQ | CCCCCCCCCCCCCCHHH | c1c-4 | uniq | 0.092 | 0.085 | 0.23 | boundary | 0.0 | |
| 13 | fp_O | P30052 | 375 | YMINFIQKLKNLPE | HHHHHHHHHHCCCC | c3-4 | uniq | 0.021 | 0.035 | 0.09 | ORD | 0.0 | |
| 14 | fp_O | P30052 | 387 | PERYMMNSVLENFTILQ | CCHHHCCCCCCCCEEEE | c1c-4 | multi | 0.03 | 0.046 | 0.127 | ORD | 0.0 | |
| 15 | fp_O | P30052 | 388 | ERYMMNSVLENFTILQ | CHHHCCCCCCCCEEEE | c1a-4 | multi | 0.029 | 0.044 | 0.129 | ORD | 0.0 | |
| 16 | fp_O | P30052 | 388 | ERYMMNSVLENFTILQ | CHHHCCCCCCCCEEEE | c1d-4 | multi | 0.029 | 0.044 | 0.129 | ORD | 0.0 | |
| 17 | fp_O | P30052 | 391 | MMNSVLENFTILQVMR | HCCCCCCCCEEEEEEE | c1a-5 | multi-selected | 0.024 | 0.031 | 0.128 | ORD | 0.286 | |
| 18 | fp_O | P30052 | 392 | MNSVLENFTILQVMR | CCCCCCCCEEEEEEE | c1b-4 | multi | 0.021 | 0.029 | 0.127 | ORD | 0.429 | |
| 19 | fp_beta_D | P30052 | 409 | ETQETLLCIAYVFEVAA | CCCCEEEEEEEEEEEEC | c1c-AT-4 | multi-selected | 0.041 | 0.036 | 0.052 | boundary | 1.0 | |
| 20 | fp_beta_D | P30052 | 410 | TQETLLCIAYVFEVAA | CCCEEEEEEEEEEEEC | c1a-AT-4 | multi-selected | 0.042 | 0.033 | 0.052 | boundary | 1.0 | |
| 21 | fp_beta_D | P30052 | 410 | TQETLLCIAYVFEVAA | CCCEEEEEEEEEEEEC | c1d-4 | multi | 0.042 | 0.033 | 0.052 | boundary | 1.0 | |
| 22 | fp_beta_D | P30052 | 411 | QETLLCIAYVFEVAA | CCEEEEEEEEEEEEC | c1b-AT-4 | multi | 0.044 | 0.031 | 0.053 | boundary | 1.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment