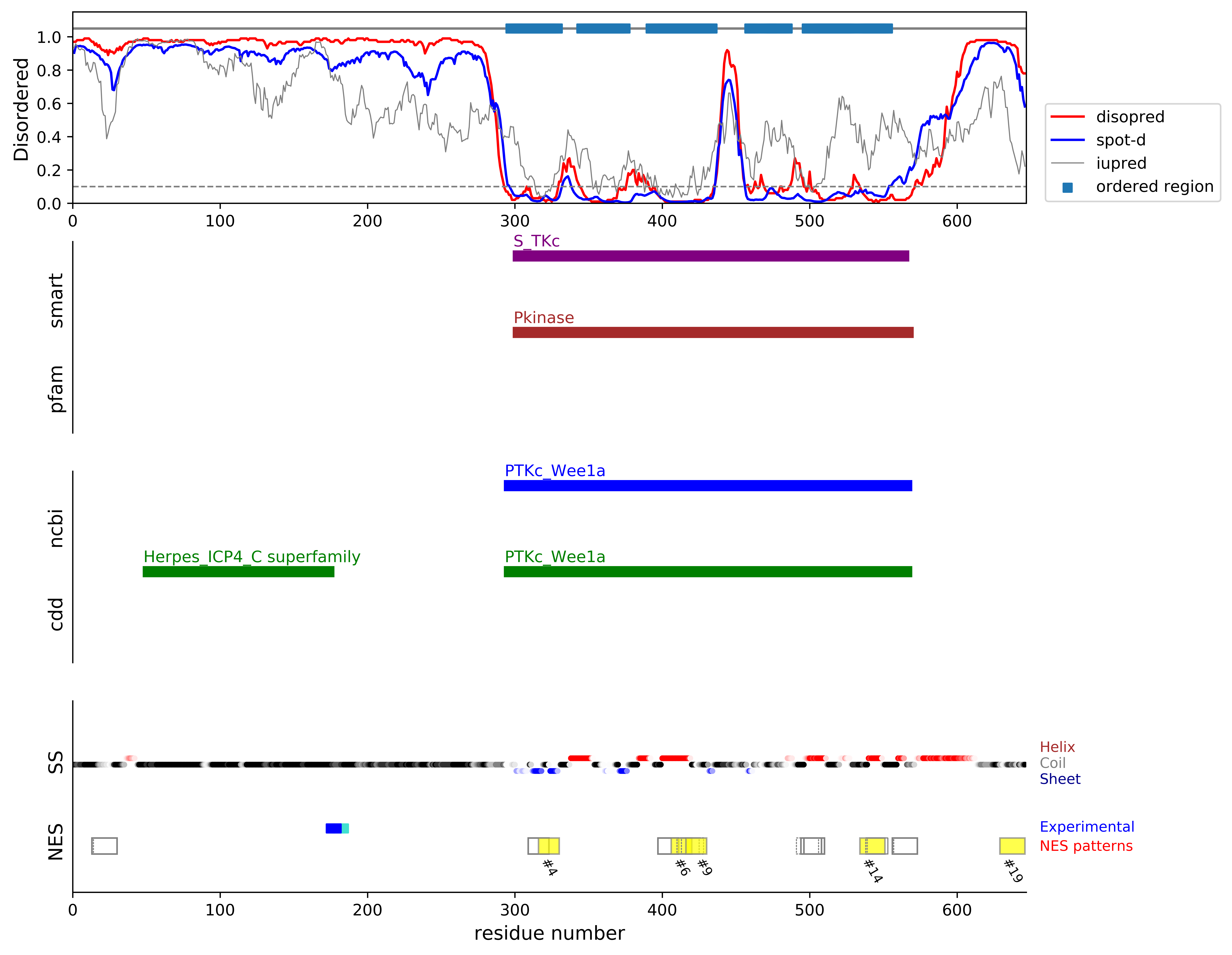

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | P30291 | 13 | RRAGAACTLRQKLIFSP | CCCCCCCCCCCCCCCCC | c1c-AT-4 | multi-selected | 0.928 | 0.833 | 0.614 | DISO | boundary|Herpes_ICP4_C superfamily; | 0.0 |
| 2 | fp_D | P30291 | 14 | RAGAACTLRQKLIFSP | CCCCCCCCCCCCCCCC | c1d-AT-4 | multi | 0.926 | 0.828 | 0.602 | DISO | boundary|Herpes_ICP4_C superfamily; | 0.0 |
| 3 | fp_beta_D | P30291 | 309 | EFGSVFKCVKRLDG | CCCEEEEEEECCCC | c3-4 | uniq | 0.036 | 0.025 | 0.09 | boundary | boundary|PTKc_Wee1a; | 1.0 |
| 4 | fp_D | P30291 | 316 | CVKRLDGCIYAIKR | EEECCCCCEEEEEE | c3-4 | uniq | 0.027 | 0.026 | 0.092 | boundary | MID|PTKc_Wee1a; | 0.286 |
| 5 | fp_D | P30291 | 397 | YFKEAELKDLLLQV | CCCHHHHHHHHHHH | c2-AT-5 | uniq | 0.019 | 0.021 | 0.068 | boundary | MID|PTKc_Wee1a; | 0.0 |
| 6 | fp_D | P30291 | 406 | LLLQVGRGLRYIHS | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.01 | 0.009 | 0.072 | boundary | MID|PTKc_Wee1a; | 0.0 |
| 7 | fp_D | P30291 | 410 | VGRGLRYIHSMSLVH | HHHHHHHHHHCCCCC | c1b-4 | multi | 0.012 | 0.01 | 0.093 | boundary | MID|PTKc_Wee1a; | 0.0 |
| 8 | fp_D | P30291 | 413 | GLRYIHSMSLVHMDI | HHHHHHHCCCCCCCC | c1b-5 | multi | 0.013 | 0.01 | 0.098 | boundary | MID|PTKc_Wee1a; | 0.0 |
| 9 | fp_D | P30291 | 416 | YIHSMSLVHMDIKP | HHHHCCCCCCCCCC | c2-5 | multi-selected | 0.014 | 0.012 | 0.12 | boundary | MID|PTKc_Wee1a; | 0.0 |
| 10 | fp_D | P30291 | 491 | NYTHLPKADIFALAL | CCCCCCHHHHHHHHH | c1b-AT-4 | multi | 0.133 | 0.025 | 0.121 | boundary | MID|PTKc_Wee1a; | 0.0 |
| 11 | fp_D | P30291 | 494 | HLPKADIFALALTV | CCCHHHHHHHHHHH | c2-AT-5 | multi-selected | 0.098 | 0.017 | 0.115 | boundary | MID|PTKc_Wee1a; | 0.0 |
| 12 | fp_D | P30291 | 494 | HLPKADIFALALTVVC | CCCHHHHHHHHHHHHH | c1d-AT-5 | multi | 0.089 | 0.017 | 0.117 | boundary | MID|PTKc_Wee1a; | 0.0 |
| 13 | fp_D | P30291 | 496 | PKADIFALALTVVC | CHHHHHHHHHHHHH | c2-AT-4 | multi-selected | 0.078 | 0.014 | 0.108 | boundary | MID|PTKc_Wee1a; | 0.0 |
| 14 | fp_D | P30291 | 534 | IPQVLSQEFTELLKVMI | CCCCCCHHHHHHHHHHC | c1c-4 | multi-selected | 0.032 | 0.058 | 0.363 | boundary | boundary|PTKc_Wee1a; | 0.0 |
| 15 | fp_D | P30291 | 538 | LSQEFTELLKVMIHP | CCHHHHHHHHHHCCC | c1b-4 | multi | 0.019 | 0.053 | 0.37 | boundary | boundary|PTKc_Wee1a; | 0.0 |
| 16 | fp_D | P30291 | 539 | SQEFTELLKVMIHP | CHHHHHHHHHHCCC | c2-AT-4 | multi | 0.018 | 0.051 | 0.373 | boundary | boundary|PTKc_Wee1a; | 0.0 |
| 17 | fp_D | P30291 | 556 | RRPSAMALVKHSVLLSA | CCCCHHHHHCCCCCCCH | c1c-AT-4 | multi-selected | 0.041 | 0.167 | 0.41 | boundary | boundary|PTKc_Wee1a; | 0.0 |
| 18 | fp_D | P30291 | 557 | RPSAMALVKHSVLLSA | CCCHHHHHCCCCCCCH | c1d-4 | multi | 0.04 | 0.171 | 0.406 | boundary | boundary|PTKc_Wee1a; | 0.0 |
| 19 | fp_D | P30291 | 629 | TSRLIGKKMNRSVSLTI | CCCCCCCCCCCCCCCCC | c1c-4 | uniq | 0.921 | 0.849 | 0.429 | DISO | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment