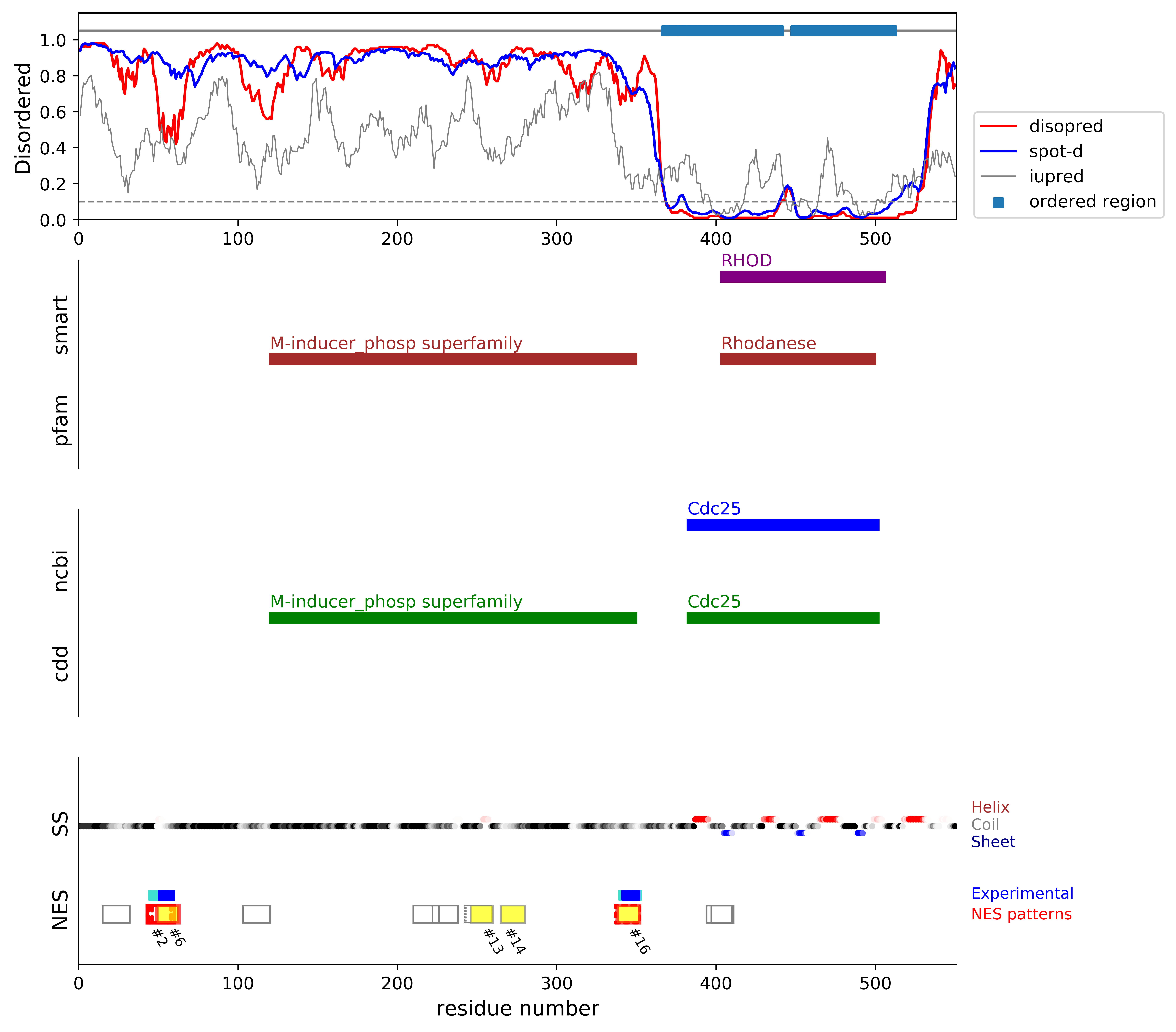

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | P30309 | 15 | TNTGLNFRTNCRMVLNL | CCCCCCCCCCCCCCCCC | c1c-AT-4 | uniq | 0.873 | 0.935 | 0.383 | DISO | 0.0 | |
| 2 | cand_D | P30309 | 43 | PEQPLTPVTDLAVGFSN ++++++*+*+* | CCCCCCCHHHCCCCCCC | c1c-AT-4 | multi-selected | 0.649 | 0.888 | 0.429 | DISO | 0.0 | |
| 3 | cand_D | P30309 | 43 | PEQPLTPVTDLAVGF ++++++*+*+* | CCCCCCCHHHCCCCC | c1b-4 | multi | 0.671 | 0.899 | 0.426 | DISO | 0.0 | |
| 4 | cand_D | P30309 | 44 | EQPLTPVTDLAVGF ++++++*+*+* | CCCCCCHHHCCCCC | c2-AT-4 | multi | 0.653 | 0.9 | 0.42 | DISO | 0.0 | |
| 5 | cand_D | P30309 | 46 | PLTPVTDLAVGFSN ++++++*+*+* | CCCCHHHCCCCCCC | c3-AT | multi | 0.594 | 0.886 | 0.421 | DISO | 0.0 | |
| 6 | cand_D | P30309 | 49 | PVTDLAVGFSNLST ++++*+*+* | CHHHCCCCCCCCCC | c3-4 | multi-selected | 0.517 | 0.861 | 0.415 | DISO | 0.0 | |
| 7 | fp_D | P30309 | 103 | QFVQFDGLFTPDLGWKA | CCCCCCCCCCCCCCCCC | c1cR-4 | uniq | 0.668 | 0.859 | 0.322 | DISO | boundary|M-inducer_phosp superfamily; | 0.0 |
| 8 | fp_D | P30309 | 210 | DPPCLDGAHDDIKMQN | CCCCCCCCCCCCCCCC | c1d-AT-4 | uniq | 0.953 | 0.911 | 0.495 | DISO | MID|M-inducer_phosp superfamily; | 0.0 |
| 9 | fp_D | P30309 | 222 | KMQNLDGFADFFSVDE | CCCCCCCCCCCCCCCH | c1a-AT-5 | multi-selected | 0.924 | 0.861 | 0.452 | DISO | MID|M-inducer_phosp superfamily; | 0.0 |
| 10 | fp_D | P30309 | 222 | KMQNLDGFADFFSVDE | CCCCCCCCCCCCCCCH | c1d-5 | multi | 0.924 | 0.861 | 0.452 | DISO | MID|M-inducer_phosp superfamily; | 0.0 |
| 11 | fp_D | P30309 | 242 | NPPGAVGNLSSSMAILL | CCCCCCCCCCCHHHHHC | c1c-AT-4 | multi | 0.87 | 0.873 | 0.548 | DISO | MID|M-inducer_phosp superfamily; | 0.0 |
| 12 | fp_D | P30309 | 243 | PPGAVGNLSSSMAILL | CCCCCCCCCCHHHHHC | c1d-4 | multi | 0.869 | 0.872 | 0.545 | DISO | MID|M-inducer_phosp superfamily; | 0.0 |
| 13 | fp_D | P30309 | 246 | AVGNLSSSMAILLS | CCCCCCCHHHHHCC | c3-4 | multi-selected | 0.861 | 0.868 | 0.483 | DISO | MID|M-inducer_phosp superfamily; | 0.0 |
| 14 | fp_D | P30309 | 265 | QDIEVSNVNNISLNR | CCCCCCCCCCCCCCC | c1b-4 | uniq | 0.911 | 0.904 | 0.402 | DISO | MID|M-inducer_phosp superfamily; | 0.0 |
| 15 | cand_D | P30309 | 337 | SLKKTLSLCDVDIST ++*++*+*+ | CCCCCCCCCCCCCCC | c1b-AT-5 | multi | 0.715 | 0.77 | 0.284 | DISO | boundary|M-inducer_phosp superfamily; | 0.0 |
| 16 | cand_D | P30309 | 338 | LKKTLSLCDVDIST ++*++*+*+ | CCCCCCCCCCCCCC | c2-4 | multi-selected | 0.709 | 0.762 | 0.276 | DISO | boundary|M-inducer_phosp superfamily; | 0.0 |
| 17 | fp_O | P30309 | 394 | IHGDFSSLVEKIFIIDC | HHCCCCCCCCCEEEEEE | c1c-4 | multi-selected | 0.014 | 0.027 | 0.058 | ORD | boundary|Cdc25; | 0.125 |
| 18 | fp_O | P30309 | 394 | IHGDFSSLVEKIFIID | HHCCCCCCCCCEEEEE | c1a-4 | multi-selected | 0.014 | 0.028 | 0.058 | ORD | boundary|Cdc25; | 0.0 |
| 19 | fp_O | P30309 | 394 | IHGDFSSLVEKIFIID | HHCCCCCCCCCEEEEE | c1d-4 | multi | 0.014 | 0.028 | 0.058 | ORD | boundary|Cdc25; | 0.0 |
| 20 | fp_O | P30309 | 397 | DFSSLVEKIFIIDC | CCCCCCCCEEEEEE | c3-4 | multi-selected | 0.014 | 0.025 | 0.043 | ORD | boundary|Cdc25; | 0.286 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment