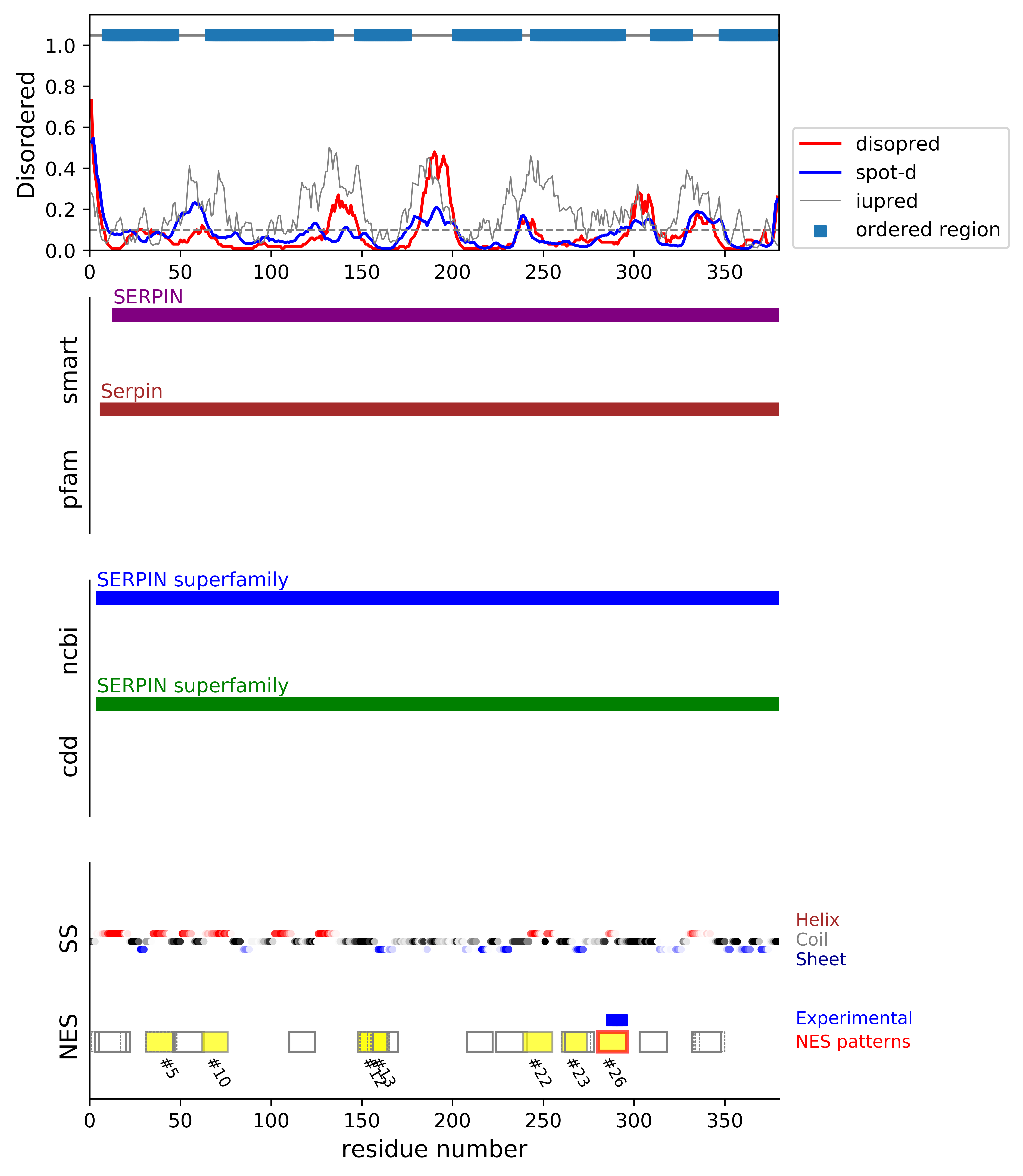

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | P30740 | 1 | XMEQLSSANTRFALDL | CCCHHHHHHHHHHHHH | c1d-AT-5-Nt | multi | 0.162 | 0.224 | 0.121 | boundary | boundary|SERPIN superfamily; | 0.0 |
| 2 | fp_D | P30740 | 3 | QLSSANTRFALDLFLAL | CHHHHHHHHHHHHHHHH | c1c-AT-5 | multi-selected | 0.086 | 0.162 | 0.102 | boundary | boundary|SERPIN superfamily; | 0.0 |
| 3 | fp_D | P30740 | 5 | SSANTRFALDLFLALSE | HHHHHHHHHHHHHHHHH | c1c-AT-4 | multi-selected | 0.052 | 0.124 | 0.083 | boundary | boundary|SERPIN superfamily; | 0.0 |
| 4 | fp_D | P30740 | 31 | SPFSISSAMAMVFLGTR | CCCCHHHHHHHHHHCCC | c1cR-4 | multi | 0.07 | 0.075 | 0.107 | boundary | MID|SERPIN superfamily; | 0.0 |
| 5 | fp_D | P30740 | 31 | SPFSISSAMAMVFLGT | CCCCHHHHHHHHHHCC | c1a-4 | multi-selected | 0.072 | 0.073 | 0.1 | boundary | MID|SERPIN superfamily; | 0.0 |
| 6 | fp_D | P30740 | 31 | SPFSISSAMAMVFLGT | CCCCHHHHHHHHHHCC | c1d-AT-4 | multi | 0.072 | 0.073 | 0.1 | boundary | MID|SERPIN superfamily; | 0.0 |
| 7 | fp_D | P30740 | 31 | SPFSISSAMAMVFLG | CCCCHHHHHHHHHHC | c1b-AT-4 | multi | 0.075 | 0.071 | 0.091 | boundary | MID|SERPIN superfamily; | 0.0 |
| 8 | fp_D | P30740 | 46 | TRGNTAAQLSKTFHFNT | CCCCCHHHHHHCCCCCC | c1c-AT-4 | multi-selected | 0.061 | 0.183 | 0.244 | boundary | MID|SERPIN superfamily; | 0.0 |
| 9 | fp_D | P30740 | 47 | RGNTAAQLSKTFHFNT | CCCCHHHHHHCCCCCC | c1d-AT-4 | multi | 0.062 | 0.188 | 0.245 | boundary | MID|SERPIN superfamily; | 0.0 |
| 10 | fp_D | P30740 | 62 | TVEEVHSRFQSLNA | CCHHHHHHHHHHHH | c3-4 | uniq | 0.059 | 0.097 | 0.255 | boundary | MID|SERPIN superfamily; | 0.0 |
| 11 | fp_D | P30740 | 110 | KTYGADLASVDFQH | HHCCCCCCCCCCCC | c2-AT-4 | uniq | 0.031 | 0.075 | 0.218 | boundary | MID|SERPIN superfamily; | 0.0 |
| 12 | fp_D | P30740 | 148 | LASGMVDNMTKLVLVNA | CCCCCCCCCCEEEEEEE | c1c-4 | multi-selected | 0.028 | 0.042 | 0.139 | boundary | MID|SERPIN superfamily; | 0.25 |
| 13 | fp_D | P30740 | 148 | LASGMVDNMTKLVLVN | CCCCCCCCCCEEEEEE | c1a-4 | multi-selected | 0.029 | 0.044 | 0.142 | boundary | MID|SERPIN superfamily; | 0.143 |
| 14 | fp_D | P30740 | 149 | ASGMVDNMTKLVLVNA | CCCCCCCCCEEEEEEE | c1a-AT-4 | multi | 0.023 | 0.041 | 0.122 | boundary | MID|SERPIN superfamily; | 0.286 |
| 15 | fp_D | P30740 | 149 | ASGMVDNMTKLVLVNA | CCCCCCCCCEEEEEEE | c1d-4 | multi | 0.023 | 0.041 | 0.122 | boundary | MID|SERPIN superfamily; | 0.286 |
| 16 | fp_D | P30740 | 149 | ASGMVDNMTKLVLVN | CCCCCCCCCEEEEEE | c1b-4 | multi | 0.023 | 0.043 | 0.123 | boundary | MID|SERPIN superfamily; | 0.286 |
| 17 | fp_beta_D | P30740 | 153 | VDNMTKLVLVNAIYFKG | CCCCCEEEEEEEEEECC | c1c-AT-4 | multi | 0.012 | 0.026 | 0.073 | boundary | MID|SERPIN superfamily; | 0.875 |
| 18 | fp_beta_D | P30740 | 155 | NMTKLVLVNAIYFKG | CCCEEEEEEEEEECC | c1b-5 | multi | 0.011 | 0.02 | 0.064 | boundary | MID|SERPIN superfamily; | 1.0 |
| 19 | fp_beta_D | P30740 | 156 | MTKLVLVNAIYFKG | CCEEEEEEEEEECC | c2-4 | multi-selected | 0.01 | 0.018 | 0.059 | boundary | MID|SERPIN superfamily; | 1.0 |
| 20 | fp_beta_D | P30740 | 208 | YIEDLKCRVLELPY | EECCCCEEEEEEEC | c3-4 | uniq | 0.012 | 0.023 | 0.095 | boundary | MID|SERPIN superfamily; | 0.571 |
| 21 | fp_beta_D | P30740 | 224 | EELSMVILLPDDIEDES | CCEEEEEECCCCCCCCC | c1cR-4 | uniq | 0.055 | 0.063 | 0.229 | boundary | MID|SERPIN superfamily; | 0.5 |
| 22 | fp_D | P30740 | 239 | ESTGLKKIEEQLTLEK | CCCCHHHHHHHCCHHH | c1d-4 | uniq | 0.088 | 0.072 | 0.35 | boundary | MID|SERPIN superfamily; | 0.0 |
| 23 | fp_D | P30740 | 260 | KPENLDFIEVNVSL | CCCCCCCEEEEEEC | c2-4 | multi-selected | 0.04 | 0.03 | 0.179 | boundary | MID|SERPIN superfamily; | 0.429 |
| 24 | fp_beta_D | P30740 | 260 | KPENLDFIEVNVSLPR | CCCCCCCEEEEEECCC | c1d-4 | multi | 0.04 | 0.029 | 0.167 | boundary | MID|SERPIN superfamily; | 0.571 |
| 25 | fp_beta_D | P30740 | 262 | ENLDFIEVNVSLPRFK | CCCCCEEEEEECCCCC | c1aR-5 | multi-selected | 0.043 | 0.027 | 0.16 | boundary | MID|SERPIN superfamily; | 0.857 |
| 26 | cand_D | P30740 | 280 | ESYTLNSDLARLGVQD * * * | CCCCCHHHHHHCCCCC | c1a-4 | uniq | 0.047 | 0.084 | 0.158 | boundary | MID|SERPIN superfamily; | 0.0 |
| 27 | fp_D | P30740 | 303 | DLSGMSGARDIFISK | CCCCCCCCCCCEEEE | c1b-AT-5 | uniq | 0.165 | 0.097 | 0.122 | boundary | MID|SERPIN superfamily; | 0.0 |
| 28 | fp_D | P30740 | 332 | EAAAATAGIATFCMLM | HHHHHHHHHHHCCCCC | c1a-AT-4 | multi-selected | 0.125 | 0.162 | 0.246 | boundary | MID|SERPIN superfamily; | 0.0 |
| 29 | fp_D | P30740 | 333 | AAAATAGIATFCMLM | HHHHHHHHHHCCCCC | c1b-AT-4 | multi | 0.126 | 0.161 | 0.238 | boundary | MID|SERPIN superfamily; | 0.0 |
| 30 | fp_D | P30740 | 334 | AAATAGIATFCMLM | HHHHHHHHHCCCCC | c2-AT-4 | multi | 0.126 | 0.16 | 0.237 | boundary | MID|SERPIN superfamily; | 0.0 |
| 31 | fp_D | P30740 | 336 | ATAGIATFCMLMPE | HHHHHHHCCCCCCC | c2-AT-4 | multi | 0.106 | 0.152 | 0.211 | boundary | MID|SERPIN superfamily; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment