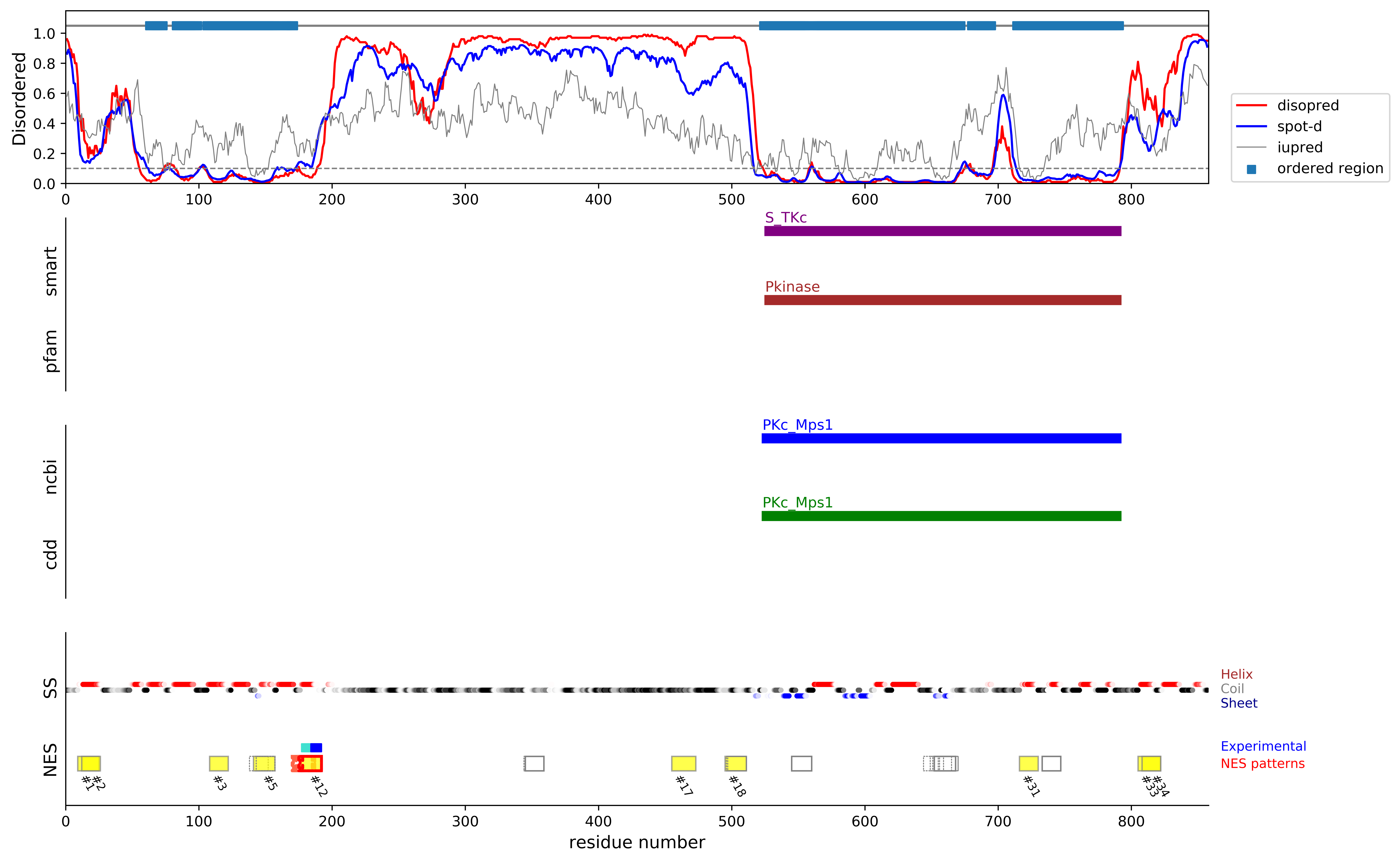

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | P33981 | 9 | RELTIDSIMNKVRDIK | CHHHHHHHHHHHHHHH | c1aR-5 | multi-selected | 0.308 | 0.188 | 0.363 | DISO | 0.0 | |
| 2 | fp_D | P33981 | 12 | TIDSIMNKVRDIKN | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.253 | 0.163 | 0.353 | DISO | 0.0 | |
| 3 | fp_D | P33981 | 108 | SFARIQVRFAELKA | HHHHHHHHHHHHHH | c3-4 | uniq | 0.018 | 0.036 | 0.253 | __ | 0.0 | |
| 4 | fp_O | P33981 | 138 | NCKKFAFVHISFAQ | CCCCCEEEHHHHHH | c2-4 | multi | 0.012 | 0.019 | 0.073 | ORD | 0.429 | |
| 5 | fp_D | P33981 | 141 | KFAFVHISFAQFELSQ | CCEEEHHHHHHHHHHC | c1a-5 | multi-selected | 0.013 | 0.022 | 0.106 | boundary | 0.143 | |
| 6 | fp_D | P33981 | 143 | AFVHISFAQFELSQ | EEEHHHHHHHHHHC | c2-5 | multi | 0.014 | 0.022 | 0.114 | boundary | 0.0 | |
| 7 | cand_D | P33981 | 170 | VERGAVPLEMLEIAL +++++ | HHCCCCCHHHHHHHH | c1b-AT-4 | multi | 0.069 | 0.119 | 0.258 | boundary | 0.0 | |
| 8 | cand_D | P33981 | 171 | ERGAVPLEMLEIALRN +++++++ | HCCCCCHHHHHHHHHH | c1a-4 | multi | 0.066 | 0.123 | 0.247 | boundary | 0.0 | |
| 9 | cand_D | P33981 | 171 | ERGAVPLEMLEIAL +++++ | HCCCCCHHHHHHHH | c2-4 | multi | 0.069 | 0.121 | 0.253 | boundary | 0.0 | |
| 10 | cand_D | P33981 | 173 | GAVPLEMLEIALRN +++++++ | CCCCHHHHHHHHHH | c2-4 | multi | 0.063 | 0.128 | 0.238 | boundary | 0.0 | |
| 11 | cand_D | P33981 | 175 | VPLEMLEIALRNLNLQK +++++++*+* | CCHHHHHHHHHHHHHCC | c1c-AT-4 | multi | 0.071 | 0.175 | 0.259 | boundary | 0.0 | |
| 12 | cand_D | P33981 | 176 | PLEMLEIALRNLNLQK +++++++*+* | CHHHHHHHHHHHHHCC | c1a-5 | multi-selected | 0.069 | 0.177 | 0.264 | boundary | 0.0 | |
| 13 | cand_D | P33981 | 176 | PLEMLEIALRNLNLQK +++++++*+* | CHHHHHHHHHHHHHCC | c1d-AT-5 | multi | 0.069 | 0.177 | 0.264 | boundary | 0.0 | |
| 14 | cand_D | P33981 | 178 | EMLEIALRNLNLQK +++++++*+* | HHHHHHHHHHHHCC | c2-5 | multi | 0.068 | 0.181 | 0.264 | boundary | 0.0 | |
| 15 | fp_D | P33981 | 344 | KSSELIITDSITLKN | CCCCCCCCCCCCCCC | c1b-AT-4 | multi | 0.925 | 0.882 | 0.448 | DISO | 0.0 | |
| 16 | fp_D | P33981 | 345 | SSELIITDSITLKN | CCCCCCCCCCCCCC | c2-AT-4 | multi-selected | 0.924 | 0.879 | 0.449 | DISO | 0.0 | |
| 17 | fp_D | P33981 | 455 | SSNTLDDYMSCFRTPVVK | CCCCCCCCCCCCCCCCCC | c4-4 | uniq | 0.917 | 0.688 | 0.318 | DISO | 0.0 | |
| 18 | fp_D | P33981 | 495 | QHQILATPLQNLQVLA | CCCCCCCCCCCCCCCC | c1a-4 | multi-selected | 0.966 | 0.729 | 0.244 | boundary | boundary|PKc_Mps1; | 0.0 |
| 19 | fp_D | P33981 | 496 | HQILATPLQNLQVLA | CCCCCCCCCCCCCCC | c1b-AT-4 | multi | 0.965 | 0.725 | 0.239 | boundary | boundary|PKc_Mps1; | 0.0 |
| 20 | fp_D | P33981 | 497 | QILATPLQNLQVLA | CCCCCCCCCCCCCC | c2-AT-5 | multi | 0.965 | 0.72 | 0.234 | boundary | boundary|PKc_Mps1; | 0.0 |
| 21 | fp_beta_O | P33981 | 545 | EKKQIYAIKYVNLEE | CCCCEEEEEEEECCC | c1b-4 | uniq | 0.032 | 0.032 | 0.142 | ORD | MID|PKc_Mps1; | 1.0 |
| 22 | fp_D | P33981 | 644 | VHSDLKPANFLIVDG | CCCCCCCCCEEEECC | c1b-AT-4 | multi | 0.01 | 0.014 | 0.098 | boundary | MID|PKc_Mps1; | 0.286 |
| 23 | fp_beta_D | P33981 | 649 | KPANFLIVDGMLKLID | CCCCEEEECCCEEEEE | c1d-4 | multi | 0.009 | 0.01 | 0.066 | boundary | MID|PKc_Mps1; | 0.571 |
| 24 | fp_beta_D | P33981 | 651 | ANFLIVDGMLKLIDFGI | CCEEEECCCEEEEEECC | c1cR-4 | multi | 0.009 | 0.01 | 0.084 | boundary | MID|PKc_Mps1; | 0.625 |
| 25 | fp_beta_D | P33981 | 651 | ANFLIVDGMLKLIDFGI | CCEEEECCCEEEEEECC | c1c-4 | multi | 0.009 | 0.01 | 0.084 | boundary | MID|PKc_Mps1; | 0.625 |
| 26 | fp_beta_D | P33981 | 652 | NFLIVDGMLKLIDFGI | CEEEECCCEEEEEECC | c1a-5 | multi-selected | 0.009 | 0.01 | 0.082 | boundary | MID|PKc_Mps1; | 0.571 |
| 27 | fp_beta_D | P33981 | 652 | NFLIVDGMLKLIDFGI | CEEEECCCEEEEEECC | c1aR-4 | multi | 0.009 | 0.01 | 0.082 | boundary | MID|PKc_Mps1; | 0.571 |
| 28 | fp_beta_D | P33981 | 652 | NFLIVDGMLKLIDFGI | CEEEECCCEEEEEECC | c1d-5 | multi | 0.009 | 0.01 | 0.082 | boundary | MID|PKc_Mps1; | 0.571 |
| 29 | fp_beta_D | P33981 | 655 | IVDGMLKLIDFGIAN | EECCCEEEEEECCCC | c1b-5 | multi | 0.012 | 0.014 | 0.101 | boundary | MID|PKc_Mps1; | 0.857 |
| 30 | fp_beta_D | P33981 | 656 | VDGMLKLIDFGIAN | ECCCEEEEEECCCC | c2-4 | multi | 0.012 | 0.015 | 0.104 | boundary | MID|PKc_Mps1; | 0.857 |
| 31 | fp_D | P33981 | 716 | DVWSLGCILYYMTY | CHHHHHHHHHHHHH | c3-4 | uniq | 0.004 | 0.025 | 0.058 | boundary | MID|PKc_Mps1; | 0.0 |
| 32 | fp_O | P33981 | 733 | PFQQIINQISKLHA | CCCCCHHHHHHHHH | c3-4 | uniq | 0.012 | 0.032 | 0.2 | ORD | MID|PKc_Mps1; | 0.0 |
| 33 | fp_D | P33981 | 805 | TTEEMKYVLGQLVGLNS | CHHHHHHHHHHHHCCCC | c1c-4 | multi-selected | 0.571 | 0.303 | 0.308 | boundary | boundary|PKc_Mps1; | 0.0 |
| 34 | fp_D | P33981 | 808 | EMKYVLGQLVGLNS | HHHHHHHHHHCCCC | c3-4 | multi-selected | 0.531 | 0.291 | 0.284 | DISO | boundary|PKc_Mps1; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment