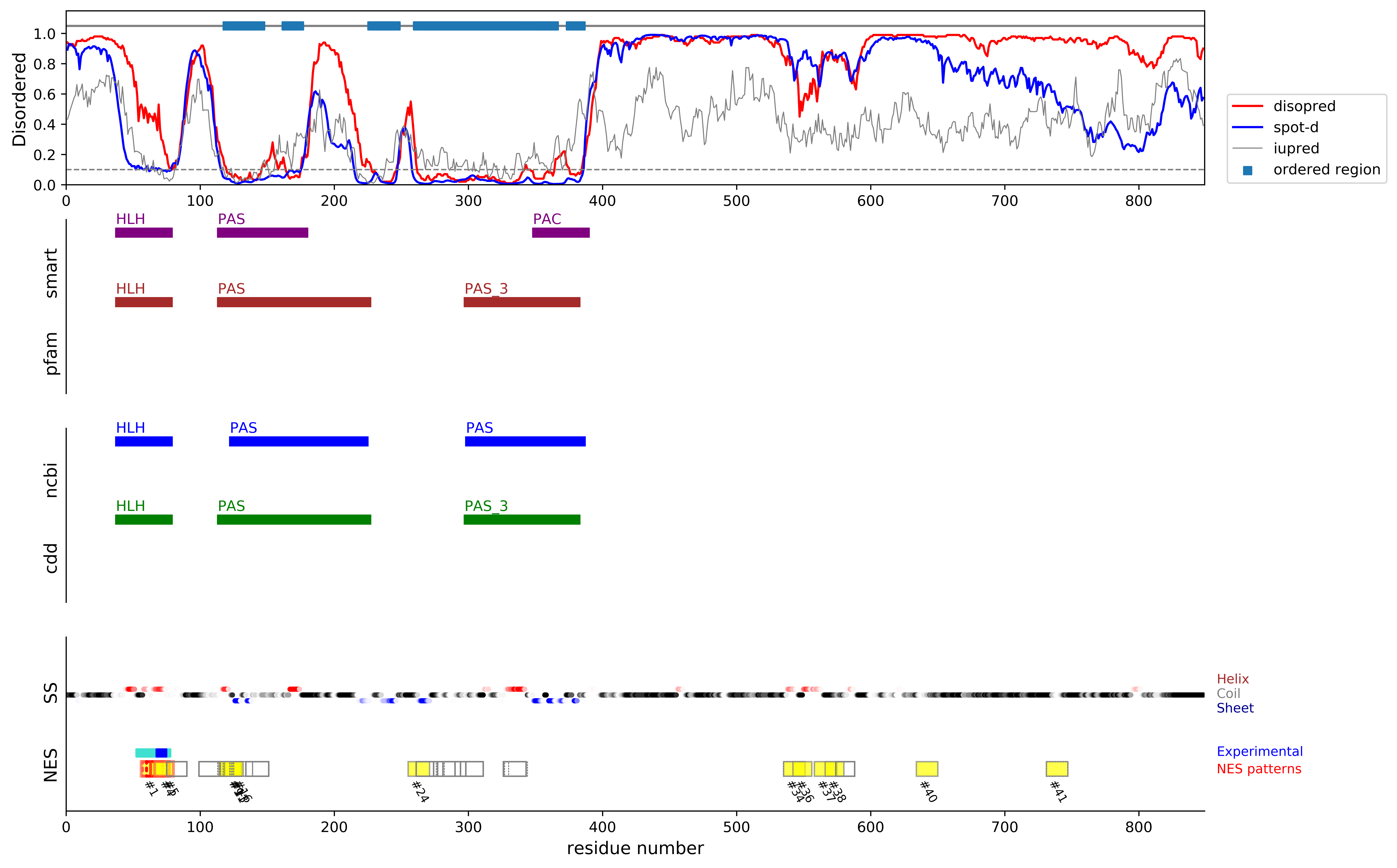

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | cand_D | P35869 | 56 | FPQDVINKLDKLSVLR ++++++++++++++*+ | CCHHHHHCHHHHHHHH | c1a-4 | multi-selected | 0.445 | 0.106 | 0.116 | DISO | small|HLH; | 0.0 |
| 2 | cand_D | P35869 | 57 | PQDVINKLDKLSVLR +++++++++++++*+ | CHHHHHCHHHHHHHH | c1b-4 | multi | 0.439 | 0.105 | 0.11 | DISO | small|HLH; | 0.0 |
| 3 | cand_D | P35869 | 60 | VINKLDKLSVLRLSV ++++++++++*+*++ | HHHCHHHHHHHHHHH | c1b-5 | multi | 0.392 | 0.104 | 0.09 | DISO | small|HLH; | 0.0 |
| 4 | cand_D | P35869 | 63 | KLDKLSVLRLSVSY +++++++*+*+++ | CHHHHHHHHHHHCC | c2-5 | multi-selected | 0.347 | 0.101 | 0.08 | DISO | small|HLH; | 0.0 |
| 5 | cand_D | P35869 | 66 | KLSVLRLSVSYLRA ++++*+*+++ | HHHHHHHHHCCCCC | c3-4 | multi-selected | 0.276 | 0.1 | 0.06 | DISO | 0.0 | |
| 6 | fp_D | P35869 | 75 | SYLRAKSFFDVALKS + | CCCCCCHHHHCCCCC | c1b-AT-4 | uniq | 0.217 | 0.21 | 0.169 | DISO | 0.0 | |
| 7 | fp_D | P35869 | 99 | DNCRAANFREGLNLQE | CCCCCCCCCCCCCCCC | c1d-AT-4 | uniq | 0.669 | 0.581 | 0.396 | boundary | boundary|PAS; | 0.0 |
| 8 | fp_D | P35869 | 113 | QEGEFLLQALNGFVLVV | CCHHHHHHHCCCEEEEE | c1c-AT-4 | multi | 0.121 | 0.054 | 0.084 | boundary | boundary|PAS; | 0.0 |
| 9 | fp_D | P35869 | 114 | EGEFLLQALNGFVLVV | CHHHHHHHCCCEEEEE | c1a-4 | multi-selected | 0.11 | 0.044 | 0.077 | boundary | boundary|PAS; | 0.0 |
| 10 | fp_D | P35869 | 114 | EGEFLLQALNGFVLVV | CHHHHHHHCCCEEEEE | c1d-AT-4 | multi | 0.11 | 0.044 | 0.077 | boundary | boundary|PAS; | 0.0 |
| 11 | fp_D | P35869 | 114 | EGEFLLQALNGFVLVVT | CHHHHHHHCCCEEEEEE | c1c-4 | multi-selected | 0.106 | 0.043 | 0.075 | boundary | boundary|PAS; | 0.125 |
| 12 | fp_D | P35869 | 115 | GEFLLQALNGFVLVV | HHHHHHHCCCEEEEE | c1b-4 | multi | 0.099 | 0.037 | 0.067 | boundary | boundary|PAS; | 0.143 |
| 13 | fp_D | P35869 | 115 | GEFLLQALNGFVLVVT | HHHHHHHCCCEEEEEE | c1aR-5 | multi-selected | 0.096 | 0.036 | 0.066 | boundary | boundary|PAS; | 0.143 |
| 14 | fp_D | P35869 | 115 | GEFLLQALNGFVLVVT | HHHHHHHCCCEEEEEE | c1d-4 | multi | 0.096 | 0.036 | 0.066 | boundary | boundary|PAS; | 0.143 |
| 15 | fp_D | P35869 | 117 | FLLQALNGFVLVVT | HHHHHCCCEEEEEE | c3-AT | multi | 0.079 | 0.026 | 0.057 | boundary | boundary|PAS; | 0.286 |
| 16 | fp_D | P35869 | 118 | LLQALNGFVLVVTT | HHHHCCCEEEEEEC | c3-4 | multi-selected | 0.068 | 0.024 | 0.051 | boundary | boundary|PAS; | 0.429 |
| 17 | fp_beta_D | P35869 | 122 | LNGFVLVVTTDALVFYA | CCCEEEEEECCCCEEEE | c1c-AT-4 | multi | 0.05 | 0.018 | 0.042 | boundary | boundary|PAS; | 0.625 |
| 18 | fp_beta_D | P35869 | 123 | NGFVLVVTTDALVFYA | CCEEEEEECCCCEEEE | c1a-AT-4 | multi | 0.049 | 0.017 | 0.039 | boundary | boundary|PAS; | 0.571 |
| 19 | fp_beta_D | P35869 | 123 | NGFVLVVTTDALVFYA | CCEEEEEECCCCEEEE | c1d-AT-4 | multi | 0.049 | 0.017 | 0.039 | boundary | boundary|PAS; | 0.571 |
| 20 | fp_D | P35869 | 124 | GFVLVVTTDALVFYA | CEEEEEECCCCEEEE | c1b-AT-5 | multi | 0.047 | 0.016 | 0.034 | boundary | boundary|PAS; | 0.429 |
| 21 | fp_D | P35869 | 125 | FVLVVTTDALVFYA | EEEEEECCCCEEEE | c2-AT-5 | multi | 0.047 | 0.016 | 0.032 | boundary | boundary|PAS; | 0.429 |
| 22 | fp_D | P35869 | 125 | FVLVVTTDALVFYA | EEEEEECCCCEEEE | c3-AT | multi | 0.047 | 0.016 | 0.032 | boundary | boundary|PAS; | 0.429 |
| 23 | fp_D | P35869 | 134 | LVFYASSTIQDYLGFQQ | CEEEECCCHHCCCCCCC | c1c-AT-5 | uniq | 0.072 | 0.03 | 0.075 | boundary | MID|PAS; | 0.125 |
| 24 | fp_D | P35869 | 255 | DGSILPPQLALFAIAT | CCCCCCCCEEEEEEEE | c1a-4 | uniq | 0.199 | 0.067 | 0.215 | boundary | boundary|PAS; | 0.429 |
| 25 | fp_beta_D | P35869 | 261 | PQLALFAIATPLQPPS | CCEEEEEEEEECCCCC | c1aR-4 | uniq | 0.053 | 0.013 | 0.168 | boundary | boundary|PAS; boundary|PAS_3; | 1.0 |
| 26 | fp_D | P35869 | 274 | PPSILEIRTKNFIFRT | CCCCCCCCCCCCCCEE | c1a-AT-4 | multi-selected | 0.056 | 0.022 | 0.167 | boundary | boundary|PAS_3; | 0.0 |
| 27 | fp_O | P35869 | 276 | SILEIRTKNFIFRT | CCCCCCCCCCCCEE | c2-AT-5 | multi | 0.058 | 0.022 | 0.168 | ORD | boundary|PAS_3; | 0.0 |
| 28 | fp_O | P35869 | 281 | RTKNFIFRTKHKLDFTP | CCCCCCCEECCCCCCCC | c1c-AT-4 | multi-selected | 0.04 | 0.026 | 0.146 | ORD | boundary|PAS_3; | 0.25 |
| 29 | fp_O | P35869 | 282 | TKNFIFRTKHKLDFTP | CCCCCCEECCCCCCCC | c1d-AT-4 | multi | 0.039 | 0.026 | 0.14 | ORD | boundary|PAS_3; | 0.286 |
| 30 | fp_O | P35869 | 294 | DFTPIGCDAKGRIVLGY | CCCCCCCCCCCCCCCCC | c1c-AT-5 | uniq | 0.035 | 0.043 | 0.117 | ORD | boundary|PAS_3; | 0.0 |
| 31 | fp_O | P35869 | 326 | HAADMLYCAESHIRMIK | CHHHHHHHHHHHHHHHH | c1c-AT-4 | multi-selected | 0.034 | 0.011 | 0.111 | ORD | MID|PAS_3; | 0.0 |
| 32 | fp_O | P35869 | 327 | AADMLYCAESHIRMIK | HHHHHHHHHHHHHHHH | c1d-AT-4 | multi | 0.035 | 0.011 | 0.114 | ORD | MID|PAS_3; | 0.0 |
| 33 | fp_O | P35869 | 330 | MLYCAESHIRMIKT | HHHHHHHHHHHHHC | c3-AT | multi | 0.046 | 0.014 | 0.121 | ORD | MID|PAS_3; | 0.0 |
| 34 | fp_D | P35869 | 535 | KNSDLYSIMKNLGIDF | CCCHHHHHHHHCCCCH | c1a-4 | multi-selected | 0.719 | 0.847 | 0.329 | DISO | 0.0 | |
| 35 | fp_D | P35869 | 535 | KNSDLYSIMKNLGIDF | CCCHHHHHHHHCCCCH | c1d-4 | multi | 0.719 | 0.847 | 0.329 | DISO | 0.0 | |
| 36 | fp_D | P35869 | 542 | IMKNLGIDFEDIRH | HHHHCCCCHHHHHH | c3-4 | uniq | 0.624 | 0.817 | 0.322 | DISO | 0.0 | |
| 37 | fp_D | P35869 | 558 | NEKFFRNDFSGEVDFRD | HHHHHHCCCCCCCCCCC | c1c-4 | uniq | 0.791 | 0.808 | 0.354 | DISO | 0.0 | |
| 38 | fp_D | P35869 | 566 | FSGEVDFRDIDLTD | CCCCCCCCCCCCCC | c2-4 | uniq | 0.845 | 0.857 | 0.311 | DISO | 0.0 | |
| 39 | fp_D | P35869 | 574 | DIDLTDEILTYVQD | CCCCCCCCCHHHHH | c3-AT | uniq | 0.796 | 0.81 | 0.295 | DISO | 0.0 | |
| 40 | fp_D | P35869 | 634 | PQQQLCQKMKHMQVNG | CCCCCCCCCCCCCCCC | c1a-4 | uniq | 0.983 | 0.938 | 0.381 | DISO | 0.0 | |
| 41 | fp_D | P35869 | 731 | TTSSLEDFVTCLQLPE | CCCCCCCCCCCCCCCC | c1a-4 | multi-selected | 0.962 | 0.574 | 0.424 | DISO | 0.0 | |
| 42 | fp_D | P35869 | 731 | TTSSLEDFVTCLQLPE | CCCCCCCCCCCCCCCC | c1d-4 | multi | 0.962 | 0.574 | 0.424 | DISO | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment