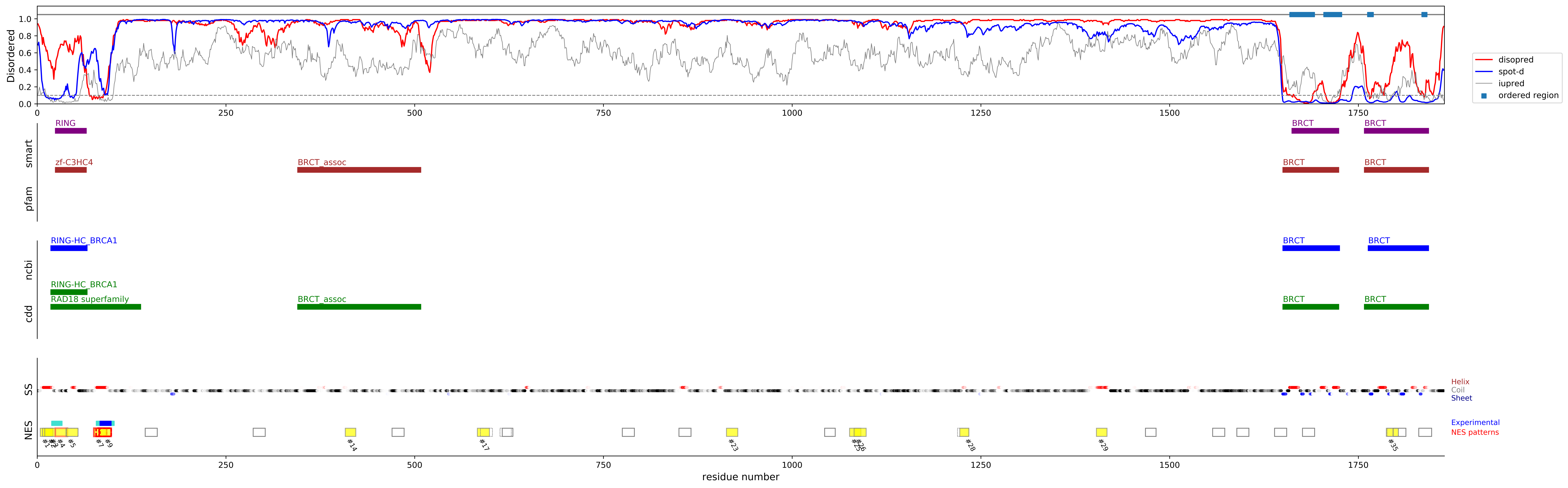

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | P38398 | 4 | SALRVEEVQNVINAMQ | CHHHHHHHHHHHHHHH | c1aR-5 | multi-selected | 0.644 | 0.158 | 0.106 | DISO | boundary|RAD18 superfamily; | 0.0 |
| 2 | fp_D | P38398 | 7 | RVEEVQNVINAMQK | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.589 | 0.096 | 0.096 | DISO | boundary|RAD18 superfamily; | 0.0 |
| 3 | fp_D | P38398 | 10 | EVQNVINAMQKILE ++ | HHHHHHHHHHHHCC | c3-4 | multi-selected | 0.511 | 0.07 | 0.071 | DISO | boundary|RAD18 superfamily; | 0.0 |
| 4 | cand_D | P38398 | 24 | CPICLELIKEPVSTKC +++++++ | CCCCCCCCCCCCCCCC | c1aR-4 | uniq | 0.646 | 0.096 | 0.03 | DISO | small|RING-HC_BRCA1; boundary|RAD18 superfamily; | 0.0 |
| 5 | fp_D | P38398 | 38 | KCDHIFCKFCMLKLLN | CCCCCCCHHHHHHHHH | c1a-4 | multi-selected | 0.675 | 0.13 | 0.027 | DISO | small|RING-HC_BRCA1; boundary|RAD18 superfamily; | 0.0 |
| 6 | fp_D | P38398 | 39 | CDHIFCKFCMLKLLN | CCCCCCHHHHHHHHH | c1b-4 | multi | 0.671 | 0.125 | 0.028 | DISO | small|RING-HC_BRCA1; MID|RAD18 superfamily; | 0.0 |
| 7 | cand_D | P38398 | 75 | ESTRFSQLVEELLKIIC +++++*+++*+ | CCHHHHHHHHHHHHHHH | c1c-4 | multi-selected | 0.088 | 0.351 | 0.13 | DISO | MID|RAD18 superfamily; | 0.0 |
| 8 | cand_D | P38398 | 78 | RFSQLVEELLKIIC +++++*+++*+ | HHHHHHHHHHHHHH | c3-4 | multi | 0.09 | 0.289 | 0.088 | DISO | MID|RAD18 superfamily; | 0.0 |
| 9 | cand_D | P38398 | 82 | LVEELLKIICAFQLDT ++++*+++*++*+*++ | HHHHHHHHHHHHCCCC | c1a-5 | multi-selected | 0.195 | 0.216 | 0.048 | DISO | MID|RAD18 superfamily; | 0.0 |
| 10 | cand_D | P38398 | 82 | LVEELLKIICAFQLDT ++++*+++*++*+*++ | HHHHHHHHHHHHCCCC | c1d-5 | multi | 0.195 | 0.216 | 0.048 | DISO | MID|RAD18 superfamily; | 0.0 |
| 11 | cand_D | P38398 | 83 | VEELLKIICAFQLDT +++*+++*++*+*++ | HHHHHHHHHHHCCCC | c1b-4 | multi | 0.202 | 0.211 | 0.046 | DISO | MID|RAD18 superfamily; | 0.0 |
| 12 | fp_D | P38398 | 143 | ENPSLQETSLSVQLSN | CCCCCCCCCCCCCCCC | c1d-AT-4 | uniq | 0.977 | 0.986 | 0.543 | DISO | boundary|RAD18 superfamily; | 0.0 |
| 13 | fp_D | P38398 | 286 | ENSSLLLTKDRMNVEK | CCCCCCCCCCCCCCCC | c1d-AT-4 | uniq | 0.899 | 0.977 | 0.559 | DISO | 0.0 | |
| 14 | fp_D | P38398 | 408 | KVADVLDVLNEVDE | HHCCCCCCCCCCCC | c3-4 | uniq | 0.986 | 0.963 | 0.46 | DISO | MID|BRCT_assoc; | 0.0 |
| 15 | fp_D | P38398 | 470 | SLPNLSHVTENLIIGA | CCCCCCCCCCCCCCCC | c1a-AT-5 | multi-selected | 0.817 | 0.944 | 0.403 | DISO | MID|BRCT_assoc; | 0.0 |
| 16 | fp_D | P38398 | 470 | SLPNLSHVTENLIIGA | CCCCCCCCCCCCCCCC | c1d-5 | multi | 0.817 | 0.944 | 0.403 | DISO | MID|BRCT_assoc; | 0.0 |
| 17 | fp_D | P38398 | 583 | KAEPISSSISNMELEL | CCCCCCCCCCCCCCCC | c1a-4 | multi-selected | 0.985 | 0.989 | 0.652 | DISO | 0.0 | |
| 18 | fp_D | P38398 | 587 | ISSSISNMELELNIHN | CCCCCCCCCCCCCCCC | c1d-4 | multi | 0.984 | 0.988 | 0.677 | DISO | 0.0 | |
| 19 | fp_D | P38398 | 613 | RKSSTRHIHALELVV | CCCCCCCCCCCEEEC | c1b-AT-4 | multi | 0.951 | 0.981 | 0.675 | DISO | 0.0 | |
| 20 | fp_D | P38398 | 616 | STRHIHALELVVSR | CCCCCCCCEEECCC | c2-AT-4 | multi-selected | 0.944 | 0.974 | 0.637 | DISO | 0.286 | |
| 21 | fp_D | P38398 | 775 | TDYGTQESISLLEVST | CCCCCCCCCCCCCCCC | c1a-AT-4 | uniq | 0.974 | 0.961 | 0.512 | DISO | 0.0 | |
| 22 | fp_D | P38398 | 850 | SELDAQYLQNTFKVSK | CCHHHHHHHHHHCCCC | c1d-AT-4 | uniq | 0.921 | 0.95 | 0.444 | DISO | 0.0 | |
| 23 | fp_D | P38398 | 913 | NESNIKPVQTVNITA | CCCCCCCCCCCCCCC | c1b-4 | uniq | 0.983 | 0.985 | 0.588 | DISO | 0.0 | |
| 24 | fp_D | P38398 | 1043 | NINEVGSSTNEVGS | CCCCCCCCCCCCCC | c3-AT | uniq | 0.988 | 0.986 | 0.611 | DISO | 0.0 | |
| 25 | fp_D | P38398 | 1076 | RGPKLNAMLRLGVLQ | CCCCCCCCCCCCCCC | c1b-4 | uniq | 0.95 | 0.97 | 0.461 | DISO | 0.0 | |
| 26 | fp_D | P38398 | 1082 | AMLRLGVLQPEVYKQS | CCCCCCCCCCCCCCCC | c1aR-4 | uniq | 0.939 | 0.953 | 0.46 | DISO | 0.0 | |
| 27 | fp_D | P38398 | 1219 | EDEELPCFQHLLFGK | CCCCCCHHHHHCCCC | c1b-4 | multi | 0.955 | 0.912 | 0.385 | DISO | 0.0 | |
| 28 | fp_D | P38398 | 1222 | ELPCFQHLLFGK | CCCHHHHHCCCC | c2-rev | multi-selected | 0.951 | 0.901 | 0.362 | DISO | 0.0 | |
| 29 | fp_D | P38398 | 1403 | NLIKLQQEMAELEA | HHHHHHHHHHHHHH | c3-4 | uniq | 0.96 | 0.824 | 0.686 | DISO | 0.0 | |
| 30 | fp_D | P38398 | 1468 | NPEGLSADKFEVSA | CCCCCCCCCCCCCC | c2-AT-4 | uniq | 0.979 | 0.826 | 0.7 | DISO | 0.0 | |
| 31 | fp_D | P38398 | 1557 | DLEGTPYLESGISLFS | CCCCCCCCCCCCCCCC | c1d-AT-5 | uniq | 0.989 | 0.894 | 0.628 | DISO | 0.0 | |
| 32 | fp_D | P38398 | 1589 | RVGNIPSSTSALKVPQ | CCCCCCCCCCCCCCCC | c1a-AT-5 | uniq | 0.99 | 0.936 | 0.606 | DISO | 0.0 | |
| 33 | fp_D | P38398 | 1639 | LTASTERVNKRMSMVV | CCCCCCCCCCEEEEEE | c1d-AT-4 | uniq | 0.604 | 0.46 | 0.554 | boundary | boundary|BRCT; | 0.143 |
| 34 | fp_D | P38398 | 1676 | LTNLITEETTHVVMKT | EECCCCCCCCEEEEEC | c1a-AT-4 | uniq | 0.033 | 0.022 | 0.367 | boundary | MID|BRCT; | 0.143 |
| 35 | fp_D | P38398 | 1787 | CGASVVKELSSFTLGT | CCCEEEECCCCCCCCC | c1a-4 | multi-selected | 0.414 | 0.077 | 0.089 | DISO | MID|BRCT; | 0.429 |
| 36 | fp_D | P38398 | 1788 | GASVVKELSSFTLGT | CCEEEECCCCCCCCC | c1b-4 | multi | 0.428 | 0.08 | 0.087 | DISO | MID|BRCT; | 0.286 |
| 37 | fp_D | P38398 | 1796 | SSFTLGTGVHPIVVVQP | CCCCCCCCCEEEEEECC | c1cR-4 | uniq | 0.642 | 0.071 | 0.184 | DISO | MID|BRCT; | 0.375 |
| 38 | fp_D | P38398 | 1830 | APVVTREWVLDSVALYQ | CCEECCCHHHHHHHHCC | c1c-AT-4 | uniq | 0.163 | 0.021 | 0.09 | DISO | boundary|BRCT; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment