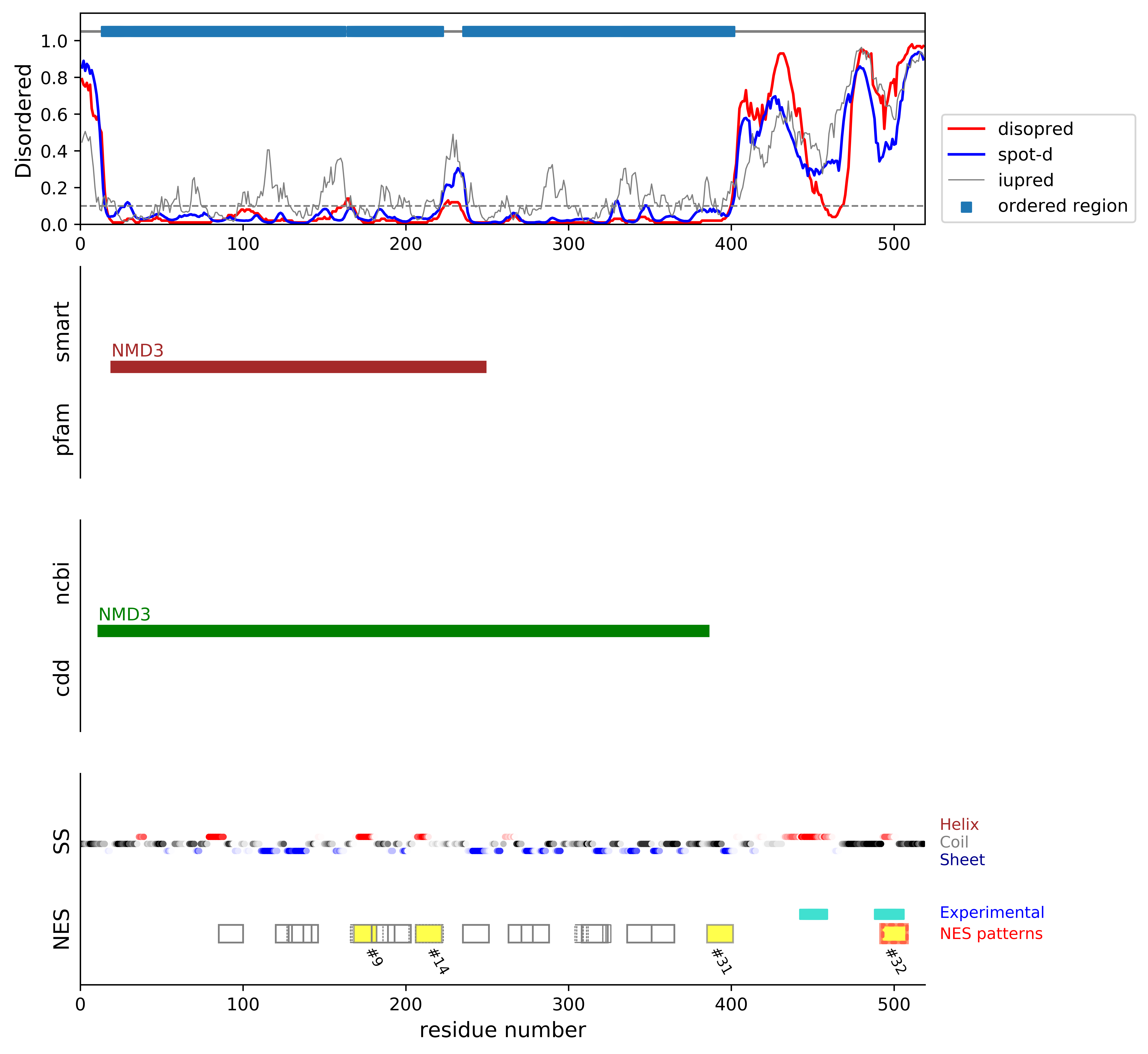

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_O | P38861 | 85 | CLRRLKGLTKVRLVD | HHHHCCCCCCEEECC | c1b-5 | uniq | 0.043 | 0.026 | 0.049 | ORD | MID|NMD3; | 0.143 |
| 2 | fp_beta_O | P38861 | 120 | GEAMTNTIIQQTFEVEY | EECCCCEEEEEEEEEEE | c1c-AT-4 | uniq | 0.017 | 0.024 | 0.121 | ORD | MID|NMD3; | 0.75 |
| 3 | fp_beta_O | P38861 | 127 | IIQQTFEVEYIVIAM | EEEEEEEEEEEEECC | c1b-AT-5 | multi | 0.013 | 0.009 | 0.072 | ORD | MID|NMD3; | 1.0 |
| 4 | fp_beta_O | P38861 | 128 | IQQTFEVEYIVIAM | EEEEEEEEEEEECC | c2-4 | multi-selected | 0.013 | 0.009 | 0.066 | ORD | MID|NMD3; | 1.0 |
| 5 | fp_beta_D | P38861 | 130 | QTFEVEYIVIAMQCPD | EEEEEEEEEECCCCCC | c1aR-4 | multi-selected | 0.014 | 0.016 | 0.067 | boundary | MID|NMD3; | 0.857 |
| 6 | fp_D | P38861 | 166 | PHKRTFLFLEQLILKH | CCCHHHHHHHHHHHHH | c1a-AT-4 | multi | 0.036 | 0.04 | 0.067 | __ | MID|NMD3; | 0.0 |
| 7 | fp_D | P38861 | 166 | PHKRTFLFLEQLILKH | CCCHHHHHHHHHHHHH | c1d-AT-4 | multi | 0.036 | 0.04 | 0.067 | __ | MID|NMD3; | 0.0 |
| 8 | fp_D | P38861 | 167 | HKRTFLFLEQLILKH | CCHHHHHHHHHHHHH | c1b-4 | multi | 0.031 | 0.038 | 0.061 | __ | MID|NMD3; | 0.0 |
| 9 | fp_D | P38861 | 168 | KRTFLFLEQLILKH | CHHHHHHHHHHHHH | c2-4 | multi-selected | 0.027 | 0.034 | 0.059 | __ | MID|NMD3; | 0.0 |
| 10 | fp_D | P38861 | 179 | LKHNAHVDTISISE | HHHCCCCCCCCEEE | c2-AT-4 | uniq | 0.021 | 0.052 | 0.117 | __ | MID|NMD3; | 0.0 |
| 11 | fp_O | P38861 | 186 | DTISISEAKDGLDFFY | CCCCEEECCCCEEEEE | c1d-AT-4 | multi | 0.017 | 0.032 | 0.134 | ORD | MID|NMD3; | 0.429 |
| 12 | fp_O | P38861 | 189 | SISEAKDGLDFFYA | CEEECCCCEEEEEC | c3-AT | multi-selected | 0.014 | 0.026 | 0.124 | ORD | MID|NMD3; | 0.429 |
| 13 | fp_D | P38861 | 206 | HAVKMIDFLNAVVPIKH | HHHHHHHHHCCCCCCCC | c1cR-4 | multi | 0.024 | 0.051 | 0.09 | boundary | MID|NMD3; | 0.0 |
| 14 | fp_D | P38861 | 206 | HAVKMIDFLNAVVPIK | HHHHHHHHHCCCCCCC | c1aR-5 | multi-selected | 0.021 | 0.046 | 0.084 | boundary | MID|NMD3; | 0.0 |
| 15 | fp_beta_D | P38861 | 235 | TGASTYKFSYSVEIVP | CCCEEEEEEEEEEEEE | c1d-AT-4 | uniq | 0.019 | 0.052 | 0.114 | boundary | MID|NMD3; | 1.0 |
| 16 | fp_O | P38861 | 263 | LAKSMGNISQFVLCS | HHHHHCCCCEEEEEE | c1b-4 | uniq | 0.025 | 0.032 | 0.068 | ORD | MID|NMD3; | 0.286 |
| 17 | fp_beta_O | P38861 | 271 | SQFVLCSKISNTVQFMD | CEEEEEEEEEEEEEEEC | c1cR-5 | multi | 0.011 | 0.012 | 0.106 | ORD | MID|NMD3; | 1.0 |
| 18 | fp_beta_O | P38861 | 271 | SQFVLCSKISNTVQFMD | CEEEEEEEEEEEEEEEC | c1c-4 | multi-selected | 0.011 | 0.012 | 0.106 | ORD | MID|NMD3; | 1.0 |
| 19 | fp_O | P38861 | 304 | APFNALADVTQLVEFIV | CCCCCCCCCCCCEEEEE | c1c-AT-4 | multi | 0.01 | 0.021 | 0.045 | ORD | MID|NMD3; | 0.0 |
| 20 | fp_O | P38861 | 305 | PFNALADVTQLVEFIV | CCCCCCCCCCCEEEEE | c1a-AT-5 | multi | 0.01 | 0.02 | 0.041 | ORD | MID|NMD3; | 0.0 |
| 21 | fp_O | P38861 | 305 | PFNALADVTQLVEFIV | CCCCCCCCCCCEEEEE | c1d-5 | multi | 0.01 | 0.02 | 0.041 | ORD | MID|NMD3; | 0.0 |
| 22 | fp_O | P38861 | 308 | ALADVTQLVEFIVLD | CCCCCCCCEEEEEEE | c1b-5 | multi | 0.01 | 0.017 | 0.042 | ORD | MID|NMD3; | 0.429 |
| 23 | fp_O | P38861 | 308 | ALADVTQLVEFIVLDV | CCCCCCCCEEEEEEEE | c1a-5 | multi-selected | 0.01 | 0.017 | 0.049 | ORD | MID|NMD3; | 0.429 |
| 24 | fp_O | P38861 | 308 | ALADVTQLVEFIVLDV | CCCCCCCCEEEEEEEE | c1d-5 | multi | 0.01 | 0.017 | 0.049 | ORD | MID|NMD3; | 0.429 |
| 25 | fp_O | P38861 | 309 | LADVTQLVEFIVLD | CCCCCCCEEEEEEE | c2-AT-4 | multi | 0.01 | 0.016 | 0.042 | ORD | MID|NMD3; | 0.429 |
| 26 | fp_beta_O | P38861 | 309 | LADVTQLVEFIVLDV | CCCCCCCEEEEEEEE | c1b-AT-4 | multi | 0.01 | 0.016 | 0.049 | ORD | MID|NMD3; | 0.571 |
| 27 | fp_beta_O | P38861 | 311 | DVTQLVEFIVLDVDS | CCCCCEEEEEEEEEE | c1b-5 | multi | 0.01 | 0.015 | 0.057 | ORD | MID|NMD3; | 0.857 |
| 28 | fp_beta_O | P38861 | 312 | VTQLVEFIVLDVDS | CCCCEEEEEEEEEE | c2-4 | multi | 0.01 | 0.014 | 0.06 | ORD | MID|NMD3; | 0.857 |
| 29 | fp_O | P38861 | 336 | ADITVARTSDLGVND | EEEEEEECCCCCCCC | c1b-AT-4 | uniq | 0.027 | 0.048 | 0.181 | ORD | MID|NMD3; | 0.429 |
| 30 | fp_O | P38861 | 351 | QVYYVRSHLGGICH | CEEEEECCCCCCCC | c3-4 | uniq | 0.011 | 0.033 | 0.121 | ORD | boundary|NMD3; | 0.429 |
| 31 | fp_D | P38861 | 385 | DGLNIDYVPDVVLVKK | CCCCCCCCCCEEEEEE | c1aR-4 | uniq | 0.034 | 0.066 | 0.139 | boundary | boundary|NMD3; | 0.143 |
| 32 | cand_D | P38861 | 492 | NIDELLDELDEMTLED ++++++++++++ | CHHHHHHHHHCCCCCC | c1a-5 | multi-selected | 0.767 | 0.512 | 0.682 | DISO | 0.0 | |
| 33 | cand_D | P38861 | 493 | IDELLDELDEMTLED +++++++++++ | HHHHHHHHHCCCCCC | c1b-4 | multi | 0.774 | 0.522 | 0.676 | DISO | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment