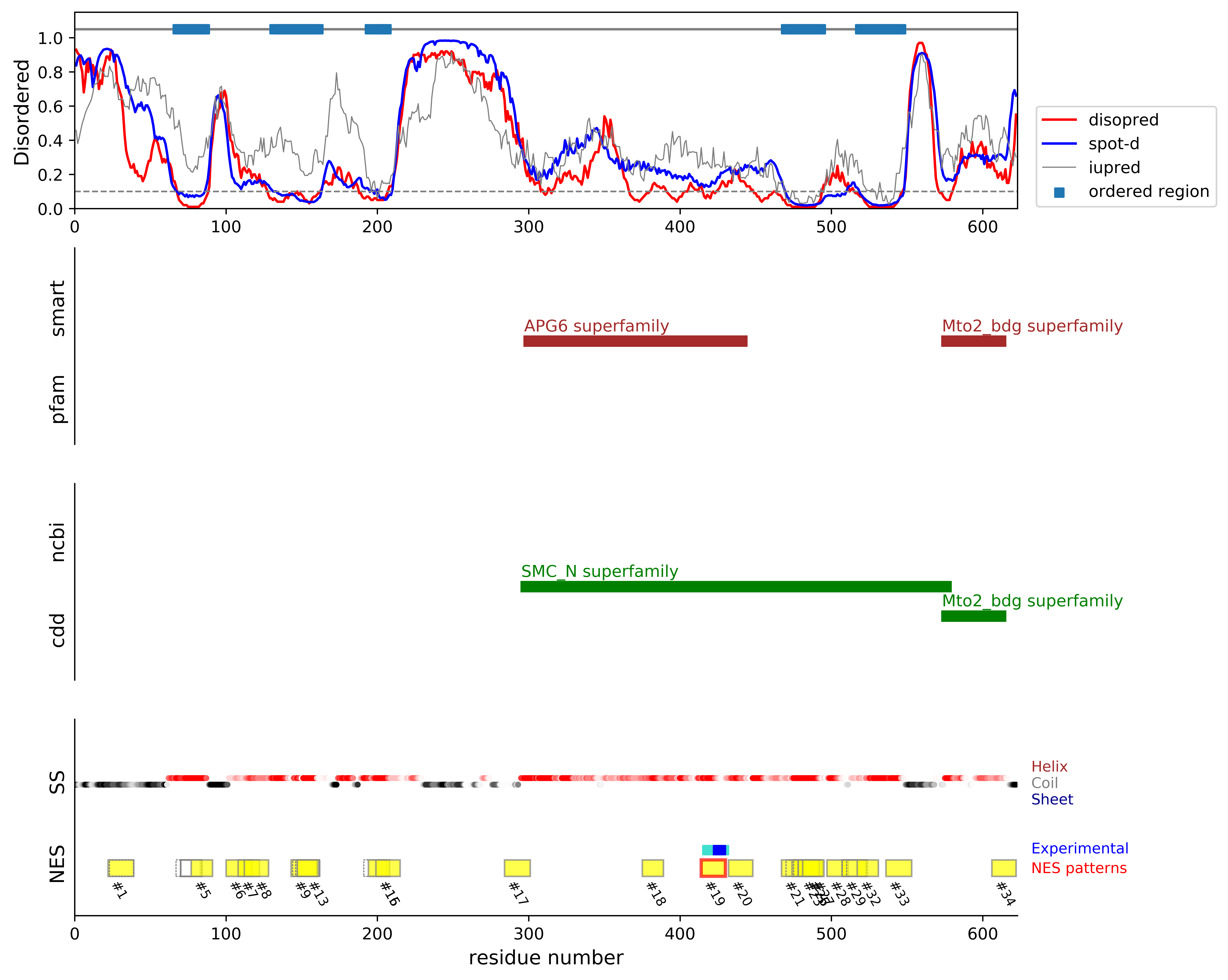

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | P39723 | 22 | DDSEFTNSMDSGMSIPS | CCCCCCCCCCCCCCCCC | c1c-4 | multi-selected | 0.619 | 0.818 | 0.688 | DISO | 0.0 | |
| 2 | fp_D | P39723 | 23 | DSEFTNSMDSGMSIPS | CCCCCCCCCCCCCCCC | c1d-AT-4 | multi | 0.602 | 0.811 | 0.683 | DISO | 0.0 | |
| 3 | fp_D | P39723 | 67 | LEKELTNAKIKIQVLY | HHHHHHHHHHHHHHHH | c1d-AT-4 | multi | 0.029 | 0.08 | 0.319 | boundary | 0.0 | |
| 4 | fp_D | P39723 | 70 | ELTNAKIKIQVLYE | HHHHHHHHHHHHHH | c3-AT | multi-selected | 0.016 | 0.077 | 0.281 | boundary | 0.0 | |
| 5 | fp_D | P39723 | 77 | KIQVLYEYIRRIPN | HHHHHHHHHHHCCC | c3-4 | uniq | 0.058 | 0.122 | 0.308 | boundary | 0.0 | |
| 6 | fp_D | P39723 | 100 | NDTDFRNSIIEGLNLEI | CCHHHHHHHHHHHHHHH | c1c-4 | uniq | 0.324 | 0.2 | 0.386 | boundary | 0.0 | |
| 7 | fp_D | P39723 | 108 | IIEGLNLEINKLKQ | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.214 | 0.146 | 0.313 | boundary | 0.0 | |
| 8 | fp_D | P39723 | 112 | LNLEINKLKQDLKAKE | HHHHHHHHHHHHHHHH | c1aR-4 | multi-selected | 0.182 | 0.154 | 0.322 | boundary | 0.0 | |
| 9 | fp_D | P39723 | 143 | NSESIVNTINHLLSFIL | HHHHHHHHHHHHHHHHH | c1c-4 | multi-selected | 0.068 | 0.052 | 0.266 | boundary | 0.0 | |
| 10 | fp_D | P39723 | 144 | SESIVNTINHLLSFIL | HHHHHHHHHHHHHHHH | c1d-4 | multi | 0.068 | 0.05 | 0.258 | boundary | 0.0 | |
| 11 | fp_D | P39723 | 146 | SIVNTINHLLSFILTH | HHHHHHHHHHHHHHHH | c1a-AT-5 | multi | 0.068 | 0.047 | 0.255 | boundary | 0.0 | |
| 12 | fp_D | P39723 | 147 | IVNTINHLLSFILTH | HHHHHHHHHHHHHHH | c1b-5 | multi | 0.067 | 0.045 | 0.251 | boundary | 0.0 | |
| 13 | fp_D | P39723 | 147 | IVNTINHLLSFILT | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.065 | 0.044 | 0.25 | boundary | 0.0 | |
| 14 | fp_D | P39723 | 191 | VLEKMDTLSKFIIQF | HHHHHHHHHHHHHHH | c1b-5 | multi | 0.057 | 0.094 | 0.16 | boundary | 0.0 | |
| 15 | fp_D | P39723 | 194 | KMDTLSKFIIQFLQ | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.06 | 0.083 | 0.135 | boundary | 0.0 | |
| 16 | fp_D | P39723 | 199 | SKFIIQFLQDFLHSKS | HHHHHHHHHHHHHHHH | c1aR-4 | multi-selected | 0.13 | 0.143 | 0.192 | boundary | 0.0 | |
| 17 | fp_D | P39723 | 284 | VESRFEKTLDTQLEIVI | CCCCCCCCCCHHHHHHH | c1c-4 | uniq | 0.418 | 0.498 | 0.328 | DISO | boundary|SMC_N superfamily; | 0.0 |
| 18 | fp_D | P39723 | 375 | NLEKLKEDIIKMKQ | HHHHHHHHHHHHHH | c3-4 | uniq | 0.109 | 0.233 | 0.247 | DISO | MID|SMC_N superfamily; | 0.0 |
| 19 | cand_D | P39723 | 414 | YINDLEKQINDLQIDK +++++++*+*++ | HHHHHHHHHHHHHHHH | c1a-5 | uniq | 0.102 | 0.16 | 0.272 | DISO | MID|SMC_N superfamily; | 0.0 |
| 20 | fp_D | P39723 | 432 | EFHVIQNQLDKLDLEN | HHHHHHHHHHHHHHHH | c1a-5 | uniq | 0.166 | 0.23 | 0.223 | DISO | MID|SMC_N superfamily; | 0.0 |
| 21 | fp_D | P39723 | 467 | YESNFIKFNQNLLLHL | HHHHHHHHHHHHHHHH | c1d-4 | multi-selected | 0.022 | 0.056 | 0.07 | boundary | MID|SMC_N superfamily; | 0.0 |
| 22 | fp_D | P39723 | 470 | NFIKFNQNLLLHLDSIF | HHHHHHHHHHHHHHHHH | c1cR-5 | multi | 0.013 | 0.033 | 0.054 | __ | MID|SMC_N superfamily; | 0.0 |
| 23 | fp_D | P39723 | 474 | FNQNLLLHLDSIFNILQ | HHHHHHHHHHHHHHHHH | c1c-4 | multi-selected | 0.011 | 0.023 | 0.055 | boundary | MID|SMC_N superfamily; | 0.0 |
| 24 | fp_D | P39723 | 475 | NQNLLLHLDSIFNILQ | HHHHHHHHHHHHHHHH | c1d-4 | multi | 0.011 | 0.022 | 0.052 | boundary | MID|SMC_N superfamily; | 0.0 |
| 25 | fp_D | P39723 | 478 | LLLHLDSIFNILQKIL | HHHHHHHHHHHHHHHH | c1aR-5 | multi-selected | 0.017 | 0.023 | 0.054 | boundary | MID|SMC_N superfamily; | 0.0 |
| 26 | fp_D | P39723 | 478 | LLLHLDSIFNILQK | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.012 | 0.021 | 0.049 | boundary | MID|SMC_N superfamily; | 0.0 |
| 27 | fp_D | P39723 | 481 | HLDSIFNILQKILQ | HHHHHHHHHHHHHH | c3-4 | multi-selected | 0.025 | 0.024 | 0.061 | boundary | MID|SMC_N superfamily; | 0.0 |
| 28 | fp_D | P39723 | 497 | SIAQFDRKMKSIKS | HHHHHHHHHHHHHC | c3-4 | uniq | 0.179 | 0.079 | 0.278 | boundary | MID|SMC_N superfamily; | 0.0 |
| 29 | fp_D | P39723 | 507 | SIKSVPNALKNLNLIQ | HHHCCHHHHHHHHHHH | c1a-5 | multi-selected | 0.109 | 0.106 | 0.253 | boundary | MID|SMC_N superfamily; | 0.0 |
| 30 | fp_D | P39723 | 507 | SIKSVPNALKNLNLIQ | HHHCCHHHHHHHHHHH | c1d-AT-5 | multi | 0.109 | 0.106 | 0.253 | boundary | MID|SMC_N superfamily; | 0.0 |
| 31 | fp_D | P39723 | 510 | SVPNALKNLNLIQP | CCHHHHHHHHHHHH | c3-AT | multi | 0.089 | 0.107 | 0.232 | boundary | MID|SMC_N superfamily; | 0.0 |
| 32 | fp_D | P39723 | 517 | NLNLIQPKLESLYT | HHHHHHHHHHHHHH | c3-4 | uniq | 0.037 | 0.055 | 0.143 | boundary | MID|SMC_N superfamily; | 0.0 |
| 33 | fp_D | P39723 | 536 | LESIINSYISSLISMET | HHHHHHHHHHHHHCCCC | c1c-4 | uniq | 0.134 | 0.132 | 0.232 | boundary | MID|SMC_N superfamily; | 0.0 |
| 34 | fp_D | P39723 | 606 | ENESLRSKLFNLSINN | HHHHHHHHHHHCCCCC | c1a-4 | uniq | 0.224 | 0.388 | 0.358 | DISO | boundary|SMC_N superfamily; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment