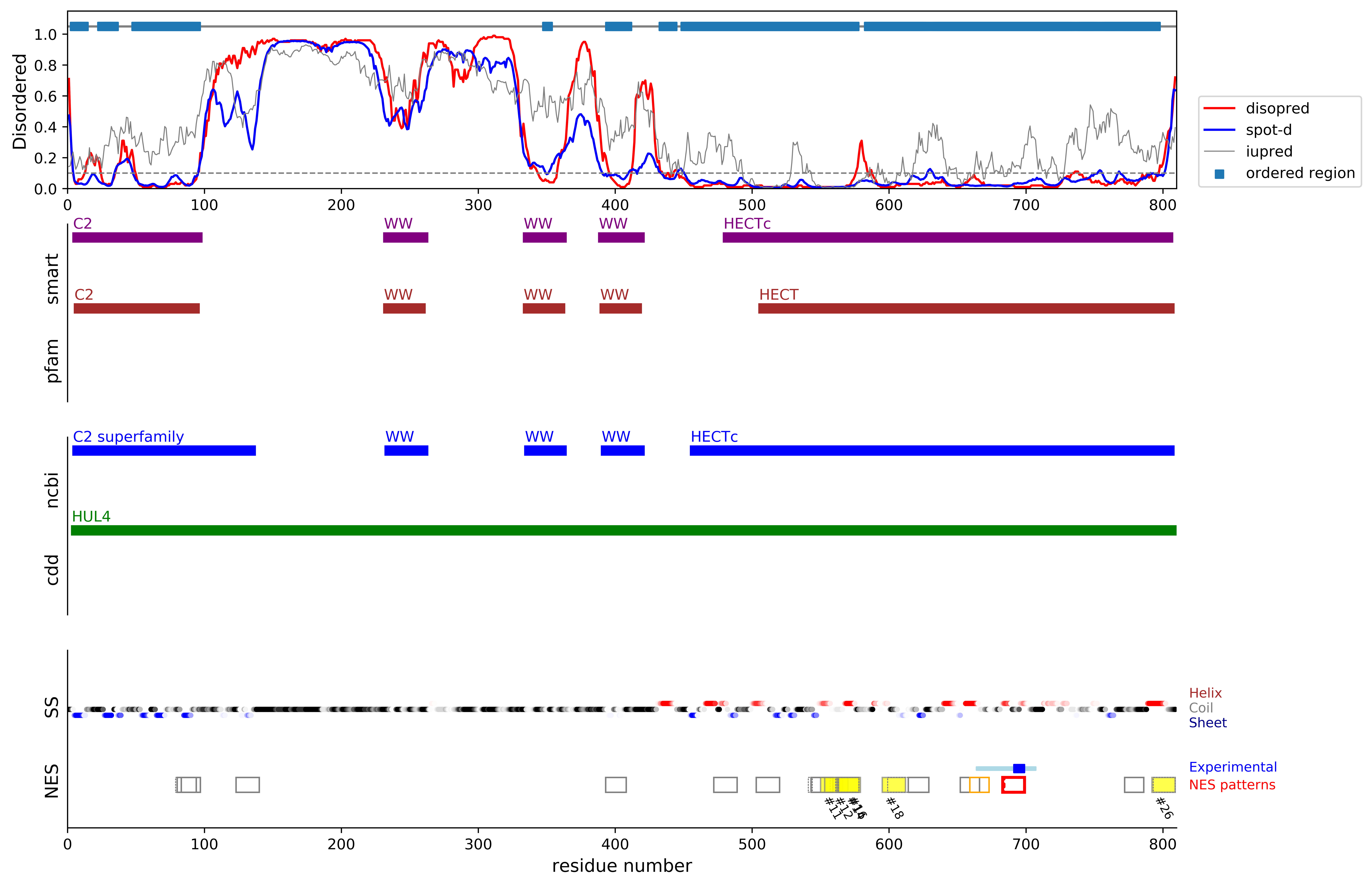

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_beta_D | P39940 | 79 | KDQGFLGVVNVRVGD | CCCCEEEEEEEECCC | c1b-4 | multi | 0.032 | 0.047 | 0.341 | boundary | MID|HUL4; | 1.0 |
| 2 | fp_beta_D | P39940 | 80 | DQGFLGVVNVRVGD | CCCEEEEEEEECCC | c2-4 | multi-selected | 0.031 | 0.044 | 0.344 | boundary | MID|HUL4; | 1.0 |
| 3 | fp_beta_D | P39940 | 83 | FLGVVNVRVGDVLG | EEEEEEEECCCCCC | c3-4 | multi-selected | 0.044 | 0.05 | 0.37 | boundary | MID|HUL4; | 0.714 |
| 4 | fp_beta_D | P39940 | 123 | DGMAVSGRLIVVLSKLP | CCCCEECCEEEECCCCC | c1cR-5 | uniq | 0.873 | 0.445 | 0.503 | DISO | MID|HUL4; | 0.75 |
| 5 | fp_D | P39940 | 393 | WEMRLTNTARVYFVD | CCEEECCCCCEECCC | c1b-AT-4 | uniq | 0.061 | 0.08 | 0.349 | boundary | MID|HUL4; | 0.286 |
| 6 | fp_O | P39940 | 472 | IMRQTPEDLKKRLMIKF | HHCCCHHHHHCEEEEEE | c1c-AT-5 | uniq | 0.022 | 0.033 | 0.263 | ORD | MID|HUL4; | 0.125 |
| 7 | fp_O | P39940 | 503 | FFFLLSHEMFNPFYCLF | HHHHHHHHHCCCCEEEE | c1cR-5 | uniq | 0.013 | 0.007 | 0.013 | ORD | MID|HUL4; | 0.0 |
| 8 | fp_O | P39940 | 541 | HLNYFKFIGRVVGLGV | CCCCEEEHHHHHHHHH | c1d-5 | multi | 0.01 | 0.01 | 0.037 | ORD | MID|HUL4; | 0.429 |
| 9 | fp_O | P39940 | 543 | NYFKFIGRVVGLGVFH | CCEEEHHHHHHHHHHC | c1a-4 | multi-selected | 0.01 | 0.008 | 0.024 | ORD | MID|HUL4; | 0.143 |
| 10 | fp_O | P39940 | 544 | YFKFIGRVVGLGVFH | CEEEHHHHHHHHHHC | c1b-5 | multi | 0.01 | 0.008 | 0.018 | ORD | MID|HUL4; | 0.0 |
| 11 | fp_D | P39940 | 550 | RVVGLGVFHRRFLDAF | HHHHHHHHCCCCCCCC | c1aR-4 | multi-selected | 0.01 | 0.008 | 0.009 | boundary | MID|HUL4; | 0.0 |
| 12 | fp_D | P39940 | 553 | GLGVFHRRFLDAFFVGA | HHHHHCCCCCCCCCHHH | c1c-5 | multi-selected | 0.011 | 0.009 | 0.008 | boundary | MID|HUL4; | 0.0 |
| 13 | fp_D | P39940 | 561 | FLDAFFVGALYKMMLRK | CCCCCCHHHHHHHHHCC | c1c-AT-5 | multi | 0.036 | 0.016 | 0.011 | boundary | MID|HUL4; | 0.0 |
| 14 | fp_D | P39940 | 562 | LDAFFVGALYKMMLRK | CCCCCHHHHHHHHHCC | c1a-4 | multi-selected | 0.038 | 0.016 | 0.011 | boundary | MID|HUL4; | 0.0 |
| 15 | fp_D | P39940 | 562 | LDAFFVGALYKMMLRK | CCCCCHHHHHHHHHCC | c1d-AT-4 | multi | 0.038 | 0.016 | 0.011 | boundary | MID|HUL4; | 0.0 |
| 16 | fp_D | P39940 | 563 | DAFFVGALYKMMLRKK | CCCCHHHHHHHHHCCC | c1aR-4 | multi-selected | 0.051 | 0.018 | 0.014 | boundary | MID|HUL4; | 0.0 |
| 17 | fp_D | P39940 | 563 | DAFFVGALYKMMLRK | CCCCHHHHHHHHHCC | c1b-4 | multi | 0.039 | 0.017 | 0.012 | boundary | MID|HUL4; | 0.0 |
| 18 | fp_D | P39940 | 595 | LNWMLENSIDGVLDLTF | HHHHHCCCCCCCEEEEE | c1c-4 | multi-selected | 0.016 | 0.03 | 0.076 | __ | MID|HUL4; | 0.0 |
| 19 | fp_D | P39940 | 599 | LENSIDGVLDLTFSA | HCCCCCCCEEEEEEE | c1b-4 | multi | 0.02 | 0.032 | 0.077 | boundary | MID|HUL4; | 0.429 |
| 20 | fp_beta_O | P39940 | 614 | DDERFGEVVTVDLKP | CCCCCCCEEEEECCC | c1b-4 | uniq | 0.03 | 0.07 | 0.204 | ORD | MID|HUL4; | 0.571 |
| 21 | fp_O | P39940 | 652 | IVDRVQEQFKAFMD . | ECHHHHHHHHHHHH | c3-4 | uniq | 0.049 | 0.021 | 0.092 | ORD | MID|HUL4; | 0.0 |
| 22 | fp_O | P39940 | 659 | QFKAFMDGFNELIP ........ | HHHHHHHCCCCCCC | c3-4 | uniq | 0.05 | 0.019 | 0.122 | ORD | MID|HUL4; | 0.0 |
| 23 | cand_O | P39940 | 683 | ELELLIGGIAEIDIED ...........*.*.. | HHHHHHHCCCCCCHHH | c1a-5 | multi-selected | 0.016 | 0.025 | 0.135 | ORD | MID|HUL4; | 0.0 |
| 24 | cand_O | P39940 | 684 | LELLIGGIAEIDIED ..........*.*.. | HHHHHHCCCCCCHHH | c1b-4 | multi | 0.015 | 0.025 | 0.135 | ORD | MID|HUL4; | 0.0 |
| 25 | fp_D | P39940 | 772 | PKSHTCFNRVDLPQ | CCCCCCCCCCCCCC | c2-AT-4 | uniq | 0.041 | 0.057 | 0.283 | boundary | MID|HUL4; | 0.0 |
| 26 | fp_D | P39940 | 792 | MKQKLTLAVEETIGFGQ | HHHHHHHHHHHCCCCCC | c1c-4 | multi-selected | 0.232 | 0.201 | 0.287 | boundary | boundary|HUL4; | 0.0 |
| 27 | fp_D | P39940 | 793 | KQKLTLAVEETIGFGQ | HHHHHHHHHHCCCCCC | c1d-AT-4 | multi | 0.242 | 0.208 | 0.287 | boundary | boundary|HUL4; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment