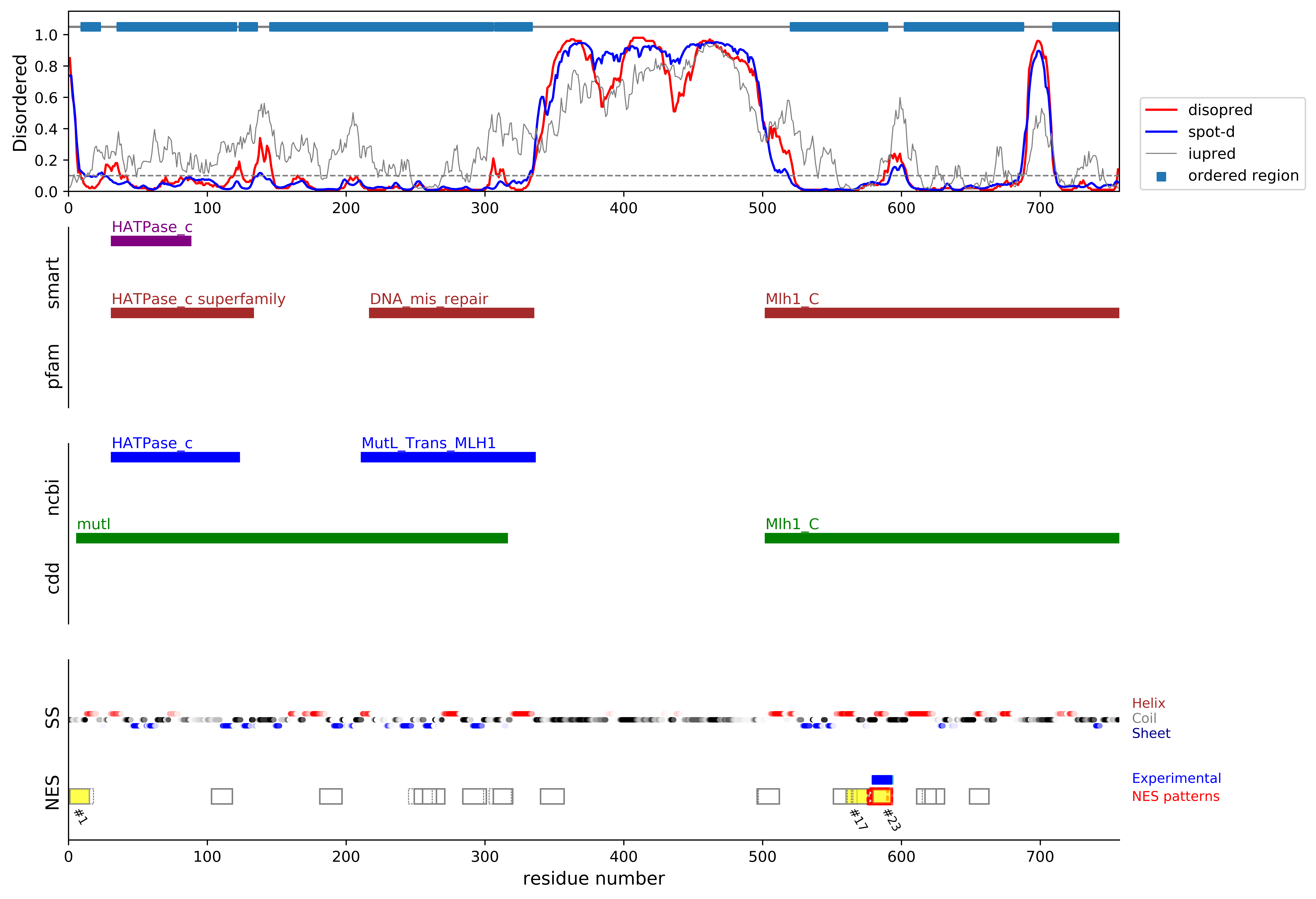

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | P40692 | 1 | XMSFVAGVIRRLDE | CCCCCCCCCCCCHH | c3-4-Nt | multi-selected | 0.284 | 0.315 | 0.078 | boundary | boundary|mutl; | 0.0 |
| 2 | fp_D | P40692 | 1 | XXMSFVAGVIRRLDETV | CCCCCCCCCCCCHHHHH | c1cR-4-Nt | multi | 0.238 | 0.276 | 0.087 | boundary | boundary|mutl; | 0.0 |
| 3 | fp_beta_D | P40692 | 103 | ALASISHVAHVTITT | CCCCCCCEEEEEEEE | c1b-5 | uniq | 0.043 | 0.031 | 0.241 | boundary | MID|mutl; | 0.571 |
| 4 | fp_O | P40692 | 181 | GRYSVHNAGISFSVKK | HHHHHCCCCCEEEEEE | c1d-AT-4 | uniq | 0.011 | 0.014 | 0.186 | ORD | MID|mutl; | 0.143 |
| 5 | fp_O | P40692 | 245 | YISNANYSVKKCIFLLF | EEECCCCCCCCCEEEEE | c1c-AT-5 | multi | 0.024 | 0.028 | 0.044 | ORD | MID|mutl; | 0.0 |
| 6 | fp_O | P40692 | 249 | ANYSVKKCIFLLFINH | CCCCCCCCEEEEEEEC | c1a-4 | multi-selected | 0.024 | 0.028 | 0.027 | ORD | MID|mutl; | 0.429 |
| 7 | fp_beta_O | P40692 | 255 | KCIFLLFINHRLVEST | CCEEEEEEECCEECCH | c1aR-4 | uniq | 0.014 | 0.015 | 0.04 | ORD | MID|mutl; | 0.714 |
| 8 | fp_beta_D | P40692 | 284 | LPKNTHPFLYLSLEI | CCCCCCCEEEEEEEE | c1b-AT-4 | multi | 0.01 | 0.014 | 0.156 | boundary | boundary|mutl; | 0.571 |
| 9 | fp_beta_D | P40692 | 284 | LPKNTHPFLYLSLEISP | CCCCCCCEEEEEEEECC | c1c-AT-4 | multi-selected | 0.01 | 0.014 | 0.163 | boundary | boundary|mutl; | 0.625 |
| 10 | fp_D | P40692 | 303 | VDVNVHPTKHEVHFLH | CCCCCCCCCCCEECCC | c1d-AT-4 | multi | 0.104 | 0.038 | 0.403 | boundary | boundary|mutl; | 0.0 |
| 11 | fp_D | P40692 | 306 | NVHPTKHEVHFLHE | CCCCCCCCEECCCH | c3-AT | multi-selected | 0.1 | 0.043 | 0.399 | boundary | boundary|mutl; | 0.286 |
| 12 | fp_D | P40692 | 340 | SRMYFTQTLLPGLAGPS | CCCCCCCCCCCCCCCCC | c1cR-4 | uniq | 0.699 | 0.651 | 0.383 | boundary | boundary|mutl; | 0.0 |
| 13 | fp_D | P40692 | 496 | PRRRIINLTSVLSLQE | CCCCCCCCHHHHHHHH | c1a-AT-4 | multi-selected | 0.433 | 0.357 | 0.387 | boundary | boundary|Mlh1_C; | 0.0 |
| 14 | fp_D | P40692 | 496 | PRRRIINLTSVLSLQE | CCCCCCCCHHHHHHHH | c1d-4 | multi | 0.433 | 0.357 | 0.387 | boundary | boundary|Mlh1_C; | 0.0 |
| 15 | fp_D | P40692 | 497 | RRRIINLTSVLSLQE | CCCCCCCHHHHHHHH | c1b-AT-4 | multi | 0.416 | 0.333 | 0.384 | boundary | boundary|Mlh1_C; | 0.0 |
| 16 | fp_O | P40692 | 551 | NTTKLSEELFYQILIYD | ECHHHHHHHHHHHHHHH | c1c-4 | uniq | 0.01 | 0.011 | 0.053 | ORD | MID|Mlh1_C; | 0.0 |
| 17 | fp_D | P40692 | 560 | FYQILIYDFANFGVLR | HHHHHHHHHCCCCEEE | c1a-4 | multi-selected | 0.017 | 0.023 | 0.024 | boundary | MID|Mlh1_C; | 0.0 |
| 18 | fp_D | P40692 | 561 | YQILIYDFANFGVLR | HHHHHHHHCCCCEEE | c1b-4 | multi | 0.017 | 0.024 | 0.023 | boundary | MID|Mlh1_C; | 0.0 |
| 19 | fp_D | P40692 | 564 | LIYDFANFGVLRLSE | HHHHHCCCCEEECCC | c1b-5 | multi | 0.025 | 0.034 | 0.025 | boundary | MID|Mlh1_C; | 0.286 |
| 20 | fp_D | P40692 | 565 | IYDFANFGVLRLSE | HHHHCCCCEEECCC | c2-AT-4 | multi | 0.026 | 0.035 | 0.026 | boundary | MID|Mlh1_C; | 0.286 |
| 21 | cand_D | P40692 | 576 | LSEPAPLFDLAMLA *++*++*+ | CCCCCCHHHHHHHH | c2-AT-4 | multi | 0.062 | 0.051 | 0.109 | boundary | MID|Mlh1_C; | 0.0 |
| 22 | cand_D | P40692 | 578 | EPAPLFDLAMLALDS *++*++*+*+ | CCCCHHHHHHHHCCC | c1b-4 | multi | 0.087 | 0.055 | 0.135 | boundary | MID|Mlh1_C; | 0.0 |
| 23 | cand_D | P40692 | 579 | PAPLFDLAMLALDS *++*++*+*+ | CCCHHHHHHHHCCC | c2-4 | multi-selected | 0.089 | 0.055 | 0.143 | boundary | MID|Mlh1_C; | 0.0 |
| 24 | fp_D | P40692 | 611 | IVEFLKKKAEMLAD | HHHHHHHHHHHHHC | c3-AT | multi-selected | 0.012 | 0.013 | 0.064 | boundary | MID|Mlh1_C; | 0.0 |
| 25 | fp_D | P40692 | 615 | LKKKAEMLADYFSLEI | HHHHHHHHHCCCCEEE | c1d-AT-4 | multi | 0.013 | 0.011 | 0.069 | boundary | MID|Mlh1_C; | 0.0 |
| 26 | fp_D | P40692 | 617 | KKAEMLADYFSLEI | HHHHHHHCCCCEEE | c2-AT-4 | multi-selected | 0.013 | 0.011 | 0.074 | boundary | MID|Mlh1_C; | 0.0 |
| 27 | fp_O | P40692 | 649 | PLEGLPIFILRLAT | CCCCHHHHHHHHCC | c2-5 | multi-selected | 0.016 | 0.025 | 0.051 | ORD | MID|Mlh1_C; | 0.0 |
| 28 | fp_O | P40692 | 649 | PLEGLPIFILRLAT | CCCCHHHHHHHHCC | c3-4 | multi-selected | 0.016 | 0.025 | 0.051 | ORD | MID|Mlh1_C; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment