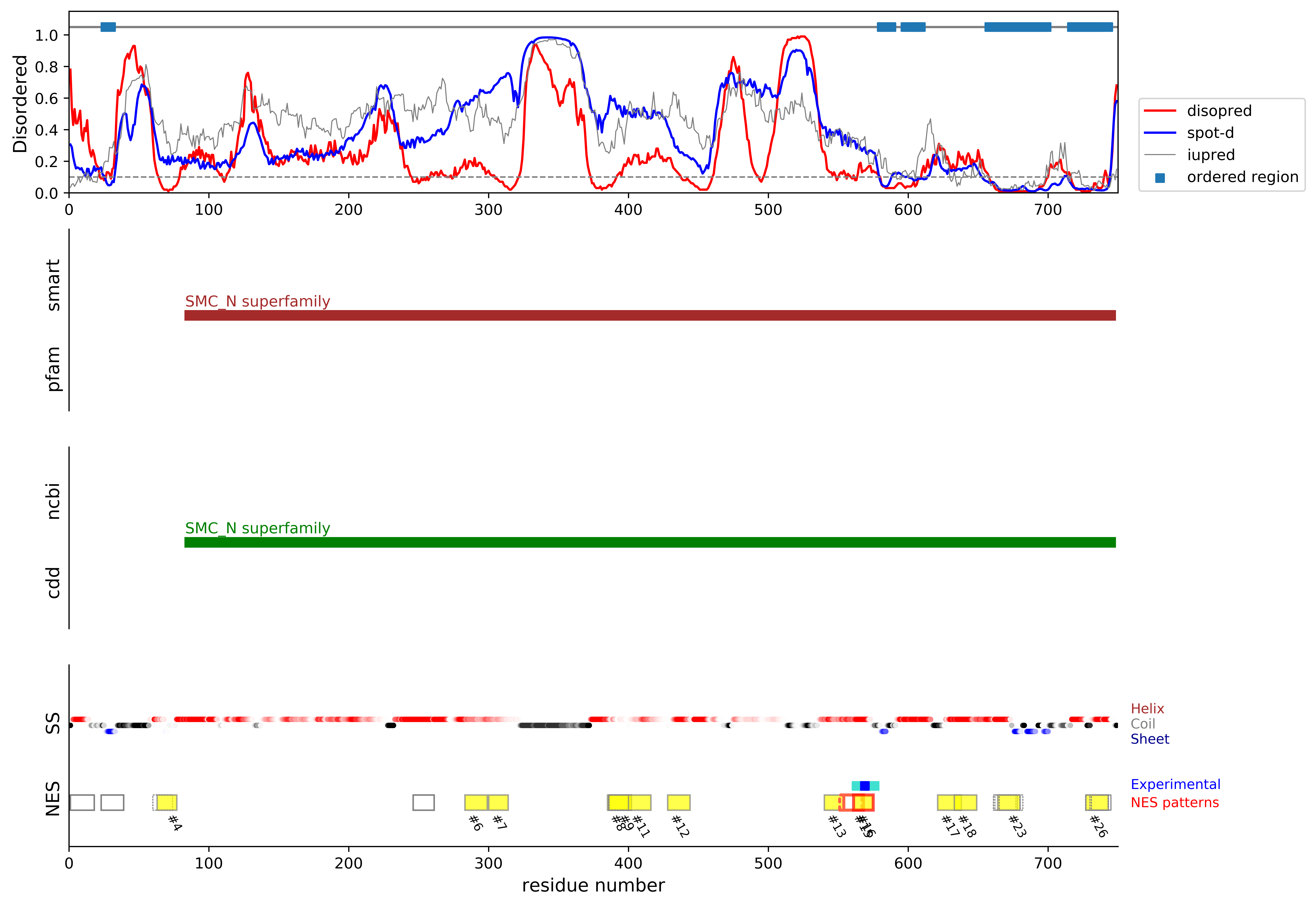

*Experimental: mutation (blue); functional region (cyan); located in the long functional sequence (lightblue)
*NES patterns: region with experimental evidence (red-orange-yellow); false positive region (gray)
| # | candidates | id | start# | sequence | secondary | class | multi-pattern | diso | spotd | iup | loc_DISO | loc_CDD | beta |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | fp_D | P40957 | 1 | XXMDVRAALQCFFSALS | CHHHHHHHHHHHHHCCC | c1cR-5-Nt | uniq | 0.424 | 0.165 | 0.08 | DISO | 0.0 | |
| 2 | fp_beta_D | P40957 | 23 | LGLEIYSIQYKMSNSG | CCCEEEEEEEECCCCC | c1aR-4 | uniq | 0.283 | 0.173 | 0.256 | DISO | 1.0 | |
| 3 | fp_D | P40957 | 60 | SNRQIQALQFKLNT | HHHHHHHHHEECCC | c2-AT-4 | multi | 0.084 | 0.257 | 0.402 | DISO | boundary|SMC_N superfamily; | 0.143 |
| 4 | fp_D | P40957 | 63 | QIQALQFKLNTLQN | HHHHHHEECCCCCC | c3-4 | multi-selected | 0.041 | 0.216 | 0.352 | DISO | boundary|SMC_N superfamily; | 0.286 |
| 5 | fp_D | P40957 | 246 | IQDQVQYTKELELAN | HHHHHHHHHHHHHHH | c1b-AT-4 | uniq | 0.087 | 0.335 | 0.538 | DISO | MID|SMC_N superfamily; | 0.0 |
| 6 | fp_D | P40957 | 283 | ENEKLQNKLSQLHVLE | HHHHHHHHHHHHHHHH | c1a-4 | uniq | 0.177 | 0.604 | 0.494 | DISO | MID|SMC_N superfamily; | 0.0 |
| 7 | fp_D | P40957 | 300 | QYENLQLENIDLKS | HHHHHHHHHHHHHH | c2-4 | uniq | 0.085 | 0.695 | 0.429 | DISO | MID|SMC_N superfamily; | 0.0 |
| 8 | fp_D | P40957 | 385 | KECLILTDMNDKLRLDN | HHHHHHHHHHHHHHHCH | c1c-4 | multi-selected | 0.116 | 0.54 | 0.378 | DISO | MID|SMC_N superfamily; | 0.0 |
| 9 | fp_D | P40957 | 386 | ECLILTDMNDKLRLDN | HHHHHHHHHHHHHHCH | c1aR-4 | multi-selected | 0.121 | 0.543 | 0.385 | DISO | MID|SMC_N superfamily; | 0.0 |
| 10 | fp_D | P40957 | 386 | ECLILTDMNDKLRLDN | HHHHHHHHHHHHHHCH | c1d-4 | multi | 0.121 | 0.543 | 0.385 | DISO | MID|SMC_N superfamily; | 0.0 |
| 11 | fp_D | P40957 | 400 | DNNNLKLLNDEMALER | CHHHHHHHHHHHHHHH | c1d-4 | uniq | 0.209 | 0.503 | 0.51 | DISO | MID|SMC_N superfamily; | 0.0 |
| 12 | fp_D | P40957 | 428 | NIVNLKRLNHELEQQK | CCHHHHHHHHHHHHHH | c1aR-4 | uniq | 0.135 | 0.297 | 0.518 | DISO | MID|SMC_N superfamily; | 0.0 |
| 13 | fp_D | P40957 | 540 | RLNELQLENVSVSR | HHHHHHHHHHHHHH | c2-5 | uniq | 0.139 | 0.377 | 0.361 | DISO | MID|SMC_N superfamily; | 0.0 |
| 14 | cand_D | P40957 | 551 | VSRELSKAQTTIQLLQ ++++ | HHHHHHHHHHHHHHHH | c1d-AT-4 | multi | 0.119 | 0.325 | 0.343 | boundary | MID|SMC_N superfamily; | 0.0 |
| 15 | cand_D | P40957 | 554 | ELSKAQTTIQLLQE +++++ | HHHHHHHHHHHHHH | c3-AT | multi-selected | 0.119 | 0.315 | 0.332 | boundary | MID|SMC_N superfamily; | 0.0 |
| 16 | cand_D | P40957 | 561 | TIQLLQEKLEKLTK ++++++*+++++ | HHHHHHHHHHHHHC | c3-4 | uniq | 0.139 | 0.282 | 0.293 | boundary | MID|SMC_N superfamily; | 0.0 |
| 17 | fp_D | P40957 | 621 | AVETVPISVYDSLNFEL | CCCCCCHHHHHHHHHHH | c1c-5 | uniq | 0.224 | 0.155 | 0.163 | boundary | MID|SMC_N superfamily; | 0.0 |
| 18 | fp_D | P40957 | 633 | LNFELKQFEQEVFKSN | HHHHHHHHHHHHHHHH | c1aR-4 | uniq | 0.205 | 0.158 | 0.102 | boundary | MID|SMC_N superfamily; | 0.0 |
| 19 | fp_D | P40957 | 661 | KSLEFIDVVNSLLGFKL | HHHHHHHHHHHHHCEEE | c1c-4 | multi | 0.015 | 0.025 | 0.034 | boundary | MID|SMC_N superfamily; | 0.0 |
| 20 | fp_D | P40957 | 661 | KSLEFIDVVNSLLGFKL | HHHHHHHHHHHHHCEEE | c1cR-4 | multi | 0.015 | 0.025 | 0.034 | boundary | MID|SMC_N superfamily; | 0.0 |
| 21 | fp_D | P40957 | 661 | KSLEFIDVVNSLLGFK | HHHHHHHHHHHHHCEE | c1aR-5 | multi | 0.015 | 0.026 | 0.033 | boundary | MID|SMC_N superfamily; | 0.0 |
| 22 | fp_D | P40957 | 662 | SLEFIDVVNSLLGFKL | HHHHHHHHHHHHCEEE | c1d-5 | multi | 0.013 | 0.023 | 0.033 | boundary | MID|SMC_N superfamily; | 0.0 |
| 23 | fp_D | P40957 | 664 | EFIDVVNSLLGFKLEF | HHHHHHHHHHCEEEEE | c1a-5 | multi-selected | 0.011 | 0.019 | 0.031 | boundary | MID|SMC_N superfamily; | 0.0 |
| 24 | fp_D | P40957 | 665 | FIDVVNSLLGFKLEF | HHHHHHHHHCEEEEE | c1b-5 | multi | 0.01 | 0.018 | 0.031 | boundary | MID|SMC_N superfamily; | 0.143 |
| 25 | fp_D | P40957 | 665 | FIDVVNSLLGFKLEFQQ | HHHHHHHHHCEEEEEEE | c1c-5 | multi | 0.01 | 0.02 | 0.03 | boundary | MID|SMC_N superfamily; | 0.25 |
| 26 | fp_D | P40957 | 727 | DRGQLPCFLATITLRL | HCCCHHHHHHHHHHHH | c1a-4 | multi-selected | 0.045 | 0.02 | 0.045 | boundary | boundary|SMC_N superfamily; | 0.0 |
| 27 | fp_D | P40957 | 727 | DRGQLPCFLATITLRL | HCCCHHHHHHHHHHHH | c1d-4 | multi | 0.045 | 0.02 | 0.045 | boundary | boundary|SMC_N superfamily; | 0.0 |
| 28 | fp_D | P40957 | 730 | QLPCFLATITLRLWE | CHHHHHHHHHHHHHH | c1b-AT-5 | multi | 0.057 | 0.024 | 0.043 | boundary | boundary|SMC_N superfamily; | 0.0 |
| 29 | fp_D | P40957 | 731 | LPCFLATITLRLWE | HHHHHHHHHHHHHH | c2-AT-4 | multi | 0.06 | 0.024 | 0.045 | boundary | boundary|SMC_N superfamily; | 0.0 |
*candidates: NES candidates and false positives annotated with "cand" and "fp", respectively;
if the segment is located in the disordered or boundary region, flagged with "D"; if the segment is located in the ordered region, flagged with "O";
if the segment's beta-strand content is over 0.5, flagged with "beta".
*sequence: Hydrophobic positions are colored in red. The positions with the experimental evidence is marked with '*' (mutation) and '+' (functional sequence in NESdb or sites in validNES).
The positions with '.' are for the region annotated in the long (more than 25 residues) functional sequence or site.
*multi-pattern: the consensus pattern is unique or multiple within the region (if the start# difference is less than 5, the segments are considered to be the same region)
*diso: average DISOPRED3-predicted disorder propensity for the segment
*spotd: average SPOT-Disorder-predicted disorder propensity for the segment
*iup: average IUPRED2A-predicted disorder propensity for the segment
*loc_DISO: location of the segment with respect to the ordered/disordered regions
*loc_CDD: location of the segment with respect to the conserved region annotated in the Conserved Domain Database (CDD)
*beta: beta-strand content in the middle of the segment